电话:020-34821111 (50条线)
传真:020-34820098 (销售部)
020-34829878 (维修部)
邮编:511442
地址:广州市番禺区兴南大道483号华粤大厦

一、适用范围:
用于微生物鉴定及微生物群落代谢分析研究的系统。
二、硬件配置
系统主机由Omnilog主机和Microstation主机两大部分组成,
2.1 Omnilog主机参数
2.1.1 主机由可控温度培养室及程控图形捕获系统部分构成,计算机内图形捕获卡通过主机中的摄像头摄取微孔板底部的图片,由软件对每个微孔中的颜色进行数字化分析,软件自动判定结果。
2.1.2主机最大容量:25层,每层可放2块板,最大容量50块板
2.1.3主机温度范围:室温+5度~45℃
2.1.4主机温度一致性:±1.5℃
2.1.5主机循环读取时间:读25块盘架(50块PM板)约需15分钟。
2.1.6最小自动读结果时间间隔:15分钟
2.1.7电源:100~240V,50或60Hz
2.1.8主机尺寸:53×81×58cm(W×H×D)
2.1.9操作环境要求:温度18~28℃,湿度20%~80%,无冷凝水。
2.2 Microstation 主机参数
2.2.1 主机为8通道96孔板读数仪
2.2.2微孔板类型:96孔板
2.2.3检测模式:终点法,动力学,线性孔扫描
2.2.4波长范围:380-900nm;
2.2.5标配滤光片数:6个
2.2.6线形范围:0-4.0 OD
2.2.7带宽:10nm
2.2.8分辨率 :0.001 OD
3.4微生物鉴定技术参数
3.4.1 鉴定原理:碳源利用原理,利用微生物对不同碳源代谢率的差异,通过检测微生物细胞利用不同碳源进行新陈代谢过程中产生的NADH与显色物质发生反应而导致的颜色变化,与标准数据库进行比对,即可得出最终鉴定结果。
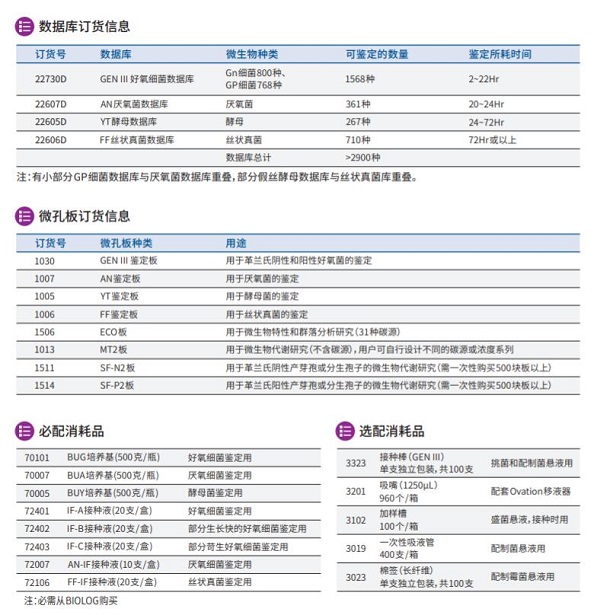
3.4.2数据库容量大于2900种(taxa),包含四大类数据库,其中Gen III好氧细菌数据库1568种(革兰氏阴性好氧菌800种,革兰氏阳性好氧菌768种),其它可选数据库有:厌氧菌361种,酵母菌267种,丝状真菌710种。几乎涵盖了所有常见的人类、动物、植物病原菌以及食品、药品和环境微生物。
3.4.3 BIOLOG丝状真菌数据库(FF),可自动鉴定包括临床、工业、农业及环境中常见青霉(153种)、曲霉(89种)、刺盘孢霉(18种)、镰刀霉(79种)、木霉及其它种等708种丝状真菌。为与传统的真菌形态学鉴定相结合,BIOLOG提供近1000多张精美的宏观和显微图片供用户参考比对,以获得更为精确的结果。
3.4.4 BIOLOG在食品、化妆品和制药工业领域提供大量的常见致病菌数据库,包括沙门氏菌(15种)、李斯特菌(7种)、大肠杆菌(8种,含阪崎肠杆菌和E.coli O157:H7)、葡萄球菌(43种好氧,2种厌氧)、弯曲菌、假单孢菌(52种)、弧菌(23种)、梭菌(60种)、志贺氏菌(4种)、耶尔森氏菌(11种)、链球菌(61种,含猪链球菌)、金黄杆菌属(32种)等,用于食品、药品及化妆品行业致病微生物鉴定、工业应用微生物研究、质量控制及产品研发等。
3.4.5 BIOLOG在植物病原菌鉴定方面,可鉴定常见的植物病原菌达到258种,如假单孢菌 (130种),伯克霍尔德菌(13种)、黄单孢菌 属(81种)、果胶杆菌属(14种)、食酸菌属 (7种)、短小杆菌(5种)、根瘤菌属(4 种)、欧文氏解淀粉菌等,数百种植物致病细菌及数百种的植物病原真菌。
3.4.6 可鉴定23种分枝杆菌(Mycobacterium)
3.4.7 可鉴定放线杆菌共22种,其中革兰氏阳性好氧放线菌8种,厌氧放线菌14种。
3.4.8 好氧细菌使用Gen III鉴定板,无需进行革兰氏染色、氧化酶和三糖铁等预测试工作,直接将培养好的纯种配制菌悬液后定量接种至鉴定板即可。
3.4.9 可生成用户自定义数据库。
3.4.10 具有进行微生物群落分析和生态研究,提供专用的ECO板,每块板31种碳源,可做3组平行。
3.4.11 鉴定时间:细菌2-22小时,酵母24-72小时,霉菌3-7天。
3.4.12 数据安全性符合美国FDA的21CFR PART11要求。
3.4.13系统鉴定步骤简单,对于好氧细菌鉴定,由机器自动扫描鉴定板,软件自动给出结果,并实现鉴定报告的直接输出,既可以形成PDF电子文本,也可直接形成打印文本。
3.5仪器配置
3.5.1 OmniLog培养箱/读数仪一套
3.5.2 OmniLog数据收集软件一套;
3.5.3 MicroLog 3数据收集软件一套;
3.5.4数据追溯管理软件2.0一套
3.5.5 MicroStation读数仪一套;
3.5.6电脑一套;
3.5.7打印机一台;
3.5.8八道电动移液器一支;
3.5.9浊度仪一台;
3.5.10 好氧菌数据库(1568种);
3.5.11 厌氧菌数据库(361种);
3.5.12 酵母菌数据库(267种);
3.5.13 丝状真菌数据库(710种);
3.5.14 安装耗材套件

更多文献请登录Biolog官网查看:https://www.biolog.com/support/bibliography/
76. New Approaches to Monitoring Cells, J.L. Fox, Bio/Technology, 1987, v.5,p. 872.
75. New Methods Aid Microbial Identification, A. Klausner, Bio/Technology, 1988, v.6, p. 756.
74. Sleuthing Out Bacterial Identities, B.R. Bochner, Nature, 1989, v.339, p. 157-158.
73. Breathprints at the Microbial Level, B.R. Bochner, ASM News, 1989, v.55, p. 536-539.
72. Efficient Ethanol Production from Glucose, Lactose, and Xylose by Recombinant Escherichia coli, F. Alterthum and L.O. Ingram, Applied and Environmental Microbiology, 1989, v.55, p. 1943-1948.
71. Pleiotropic Changes Resulting From Depletion of Era, an Essential GTP-binding Protein in Escherichia coli, C.G. Lerner and M. Inouye, Molecular Microbiology, 1991, v.5, p. 951-957.
70. The Matrix Approach: Microbial Retention Testing of Sterilizing-Grade Filters with Final Parenteral Products, Part II, R.V. Levy, K.S. Souza, D.L. Hyde, S.M. Gambale, and C.B. Neville, Pharmaceutical Technology, 1991, v.15, p. 58-68.
69. Identification of Glutaraldehyde-Resistant, Rapidly Growing Mycobacteria Based on Sole-Carbon-Source Utilization, C. Roberts, S. Guida, and H. Chan-Myers, Abstracts of the 92nd Meeting of the American Society for Microbiology, 1992, p.173.
68. Characterization of Facultative Thermophilic Bacteria Isolated From the Air at Six Fiberglass Manufacturing Plants, D. Dewitt, and P. Stukus, Abstracts of the 93rd Meeting of the American Society for Microbiology, 1993, p.411.
67. Comparison of the MIDI and BIOLOG Systems for Identification of Indoor Airborne Bacteria, L.E. Pifer, T.M. Bowen, J.R. Meldrum, and L.D. Stetzenbach, Abstracts of the 93rd Meeting of the American Society for Microbiology, 1993, p.412.
66. Identification of Environmental Bacteria, B.R. Bochner, Pharmaceutical Manufacturing International, 1993, p.223-225.
65. R & D 100 Awards: Biotechnology, R & D Magazine, October, 1991, p.44.
64. Microbes: Improved Methods for Recovering Bacteria from HPW Water, J.L. Clancy and L. Cimini, Ultrapure Water, 1991, p. 25-38.
63. Classification of Acinetobacter Strains by the Biolog System, A.T. Bernards, J. van der Toorn, B.R. Bochner, L. Dijkshoorn, Abstract Presentation of FEMS Meeting, Granada, Spain, September, 1993, p.64.
62. Gellan-related Polysaccharides and the Genus Sphingomonas, T. Pollack, Journal of General Microbiology, 1993, v.139, p. 1939-1945.
61. Denitrification under various aeration conditions in Comamonas sp., strain SGLY2, D. Patureau, J. Davison, N. Bernet, and R. Moletta, FEMS Microbiology Ecology 1994, v.14, p. 71-78.
60. Use of the Biolog-System for Rapid Characterization and Classification of Coryneform Isolates, K. Martin, I. Groth, J. Meyer, and P. Schumann, Abstracts of the 1993 FEMS Conference on Microbial Taxonomy, Granada, Spain, 1993.
59. Intraspecific Metabolic Diversity Among Strains of Burkholderia cepacia Isolated from Decayed Onions, Soils and the Clinical Environment, D.S. Yohalem, and J.W. Lorbeer, A. Van Leeuwenhoek Journal, 1994, v.65, p. 111-131.
58. Phenotypic Characterisation of Acinetobacter Strains of 13 DNA-DNA Hybridisation Groups by Means of the Biolog System, A.T. Bernards, L. Dijkshoorn, J. Van der Toorn, B.R. Bochner, and C.P.A. Van Boven, Journal of Medical Microbiology, 1995, v.42, p. 113-119.
57. Distribution of Alginate Genes in Bacterial Isolates from Corroded Metal Surfaces, W.H. Wallace, J.F. Rice, D.C. White, and G.S. Sayler, Microbial Ecology, 1994, v.27, p. 213-223.
56. Phenotypic Profiles for the Assessment and Identification of Microbial Diversity, B.R. Bochner, Abstracts of the Australian Society for Microbiology, Microbiology Australia, September, 1995, v.16, S21.4.
55. Metabolic Taxonomy of over 1100 Species of Bacteria and Yeast based on Biolog MicroPlates, B.R. Bochner, Abstracts of The Australian Society for Microbiology, Microbiology Australia, September, 1995, v.16, PO3.15.
54. The Biolog MicroStation System and General Procedures for Identifying Environmental Bacteria and Yeast, B.R. Bochner, Automated Microbial Identification and Quantitation: Technologies for the 2000s, Wayne P. Olson, ed., Interpharm Press, 1996, p.13-51.
53. Identification of Gram Negative Aerobic / Microaerophilic Rods with Four Automated and Semi-Automated Systems, M.E. Wilson, and N.E. Jackson, Abstracts and Poster of the 96th General Meeting of the American Society for Microbiology, 1996, p. 394.
52. Polyphasic Taxonomy, a Consensus Approach to Bacterial Systematics, P. VanDamme, B. Pot, M. Gillis, P. DeVos, K. Kersters, and J. Swings, Microbiological Reviews, 1996, v.60. p. 407-438.
51. Ability of Biolog and Biotype-100 Systems to Reveal Taxonomic Diversity of the Pseudomonads, P.A.D. Grimont, M. Vancanney, M. Lefevre, K. Vandemeulebroecke, L. Vauterin, R. Brosch, K. Kersters, and F. Grimont, Systematic Applied Microbiology, v.19, pp. 510-527.
50. Inhibition of Capsule Production in Bacteria by Thioglycolate, A. Franco-Buff, P. Domenico, and B. R. Bochner, Abstracts of the 98th Meeting of the American Society for Microbiology, 1998, Q-67
49. Burkholderia Caribensis sp. Nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique, Wafa Achouak, Richard Christen, Mohamed Barakat, Marie-Helene Martel, and Thierry Heulin, International Journal of Systematic Bacteriology, 1999
48. The Global Regulatory Protein FruR Modulates the Direction of Carbon Flow in Escherichia Coli, Tom M. Ramseier, Stefan Bledig, Valerie Michotey, Rita Feghali, and Milton H. Saier Jr., Molecular Microbiology, 1995
47. Pig and Goat Blood As Substitutes for Sheep Blood in Blood-Supplemented Agar Media, C. Anand, R. Gordon, H. Shaw, K. Fonseca, and M. Olsen, Journal of Clinical Microbiology, 2000, v.38, pp. 591-594.
46. Comparison of API 20NE and Biolog GN Identification Systems Assessed by Techniques of Multivariate Analyses, Jaak Truu, Ene Talpsep, Eeva Heinaru, Ulrich Stottmeister, Helmuth Wand, and Ain Heinaru, Journal of Microbiological Methods, 1999, v.36, pp. 193-201.
45. http://ijs.sgmjournals.org/content/51/1/169.short
44. Arthrobacter siderocapsulatus Dubinina and Zhdanov 1975AL is a Later Subjective Synonym of Pseudomonas putida (Trevisan 1889) Migula 1895AL, J. Chun, M. Rhee, J. Han, and K. S. Sook Bae, International Journal of Systematic and Evolutionary Microbiology, 2001, v.51, pp. 169-1
43. Taxonomic Characterization of Ochrobactrum sp. Isolates from Soil Samples and Wheat Roots, and Description of Ochrobactrum tritici sp. nov. and Ochrobactrum gringonense sp. nov., M. Lebuhn, W. Achouak, M. Schloter, O. Berge, H. Meier, M. Barakat, A. Hartmann, and T. Heulin, International Journal of Systematic and Evolutionary Microbiology, 2000, v.50, pp. 2207-2223.
42. A Comparative Evaluation of Five Typing Techniques for Determining the Diversity of Fluorescent Pseudomonads, S.L. Dawson, J.C. Fry, and B.N. Dancer, Journal of Microbiological Methods, 2002, v.50, pp. 9-22.
41. Chlorxanthomycin, a Fluorescent, Chlorinated, Pentacyclic Pyrene from a Bacillus sp., A. Magyarosy, J.Z. Ho, H. Rapoport, S. Dawson, J. Hancock, and J.D. Keasling, Applied and Environmental Microbiology, 2002, v.68, pp. 4095-4101.
40. Strain Diversity within a Microbial Collection, D. DiTullio, N. Rocheleau, M.K. Talbot, W. Millett, J. McAlpine, and P. Robakiewicz, The Journal of Antibiotics, 2002, v.55, pp. 899-903.
39. Kineococcus radiotolerans sp. nov., a Radiation-Resistant, Gram-Positive Bacterium, R.W. Phillips, J. Wiegel, C.J. Berry, C. Fliermans, A.D. Peacock, D.C. White, and L.J. Shimkets, International Journal of Systematic and Evolutionary Microbiology, 2002, v.52, pp. 933-938.
38. Aquaspirillum dispar Hylemon et al. 1973 and Microvirgula aerodenitrificans Patureau et. al. 1998 are Subjective Synonyms, I. Cleenwerck, M. De Wachter, B. Hoste, D. Janssens and J. Swings, International Journal of Systematic and Evolutionary Microbiology, 2003, v.53, pp. 1457-1459.
37. Microbial Testing in Support of Aseptic Processing, A.M. Cundell, Pharmaceutical Technology , 2004, pp. 56-66 .
36. Isolation and Partial Characterization of a Purple Pigment Producing Bacterium from the Tennessee Copper Basin, A.M.l. Gaston, A. Ejiofor, T. Johnson, Abstract from the ASM 105th General Meeting June 5th-June 9th 2005.
35. Anoxyphototroph Purple Non-Sulfur Bacteria Isolated from Water Reservoirs of Puerto Rico, K.Y. Flores-Burgos, A.J. Toro-Ramos, I.M. Almodovar-Rodgriguez, N. Figueroa-Matias, C. Rios-Velazquez, Abstract from the ASM 105th General Meeting June 5th-June 9th 2005.
34. Mycobacterium-Rhizosphere Interactions and Effects on PAH Degradation, R.D. Child, C. Miller, and A.J. Anderson, Abstract from the ASM 105th General Meeting June 5th-June 9th 2005.
33. One-to-One Comparison: A New Approach to Analyzing Metabolic Diversity Data, C. San Miguel, M. Dulinski, and R.L. Tate, III, Abstract from the 104th General Meeting of the American Society of Microbiology, May 2004.
32. Porphyromonas uenonis sp. nov., a Pathogen for Humans Distinct from P. asaccharolytica and P. endodontalis, S.M. Finegold, M-L. Vaisanen, M. Rautio, E. Eerola, P. Summanen, D. Molitoris, Y. Song, C. Liu, and H. Jousimies-Somer, Journal of Clinical Microbiology, 2004, v.42, pp. 5298-5301.
31. Proline-Based Modulation of 2,4-Diacetylphloroglucinol and Viable Cell Yields in Cultures of Pseudomonas fluorescens Wild-Type and Over-Producing Strains, P.J. Slininger and M.A. Shea-Andersh, Applied Microbiology and Biotechnology, 2005, v.68, pp. 630-638.
30. Assessment of Self-Organizing Maps to Analyze Sole-Carbon Source Utilization Profiles, J. Leflaive, R. Cereghino, M. Danger, G. Lacroix, L. Ten-Hage, Journal of Microbiological Methods, 2005, v.62, pp. 89-102.
29. Nereida ignava gen. nov., sp. nov., a Novel Aerobic Marine c-Proteobacterium that is Closely Related to Uncultured Prionitis (Alga) Gall Symbionts, M.J. Pujalte, M.C. Macian, D.R. Arahal, W. Ludwig, K.H. Schleifer, and E. Garay, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 631-636.
28. Paucibacter toxinivorans gen. nov., sp. nov., a Bacterium that Degrades Cyclic Cyanobacterial Hepatotoxins Microcystins and Nodularin, J. Rapala, K.A. Berg, C. Lyra, R. Maarit Niemi, W. Manz, S. Suomalainen, L. Paulin, and K. Lahti, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 1563-1568.
27. Characterization of Alkaliphilic Bacillus Strains Used in Industry: Proposal of Five Novel Species, Y. Nogi, H. Takami, and K. Horikoshi, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2309-2315.
26. Methylobacterium isbiliense sp. nov., Isolated from the Drinking Water System of Sevilla, Spain, V. Gallego, M.T. Garcia, and A. Ventosa, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2333-2337.
25. Thalassobius mediterraneus gen. nov., sp. nov., and Reclassification of Rugeria gelatinovorans as Thalassobius gelatinovorus comb. nov., D.R. Arahal, M.C. Macian, E. Garay, and M.J. Pujalte, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2371-2376.
24. Cyclobacterium amurskyense sp. nov., a Novel Marine Bacterium Isolated from Sea Water, O.I. Nedashkovskaya, S.B. Kim, M.S. Lee, M.S. Park, K.H. Lee, A.M. Lysenko, H.W. Oh, V.V. Mikhailov, and K.S. Bae, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2391-2394.
23. Deinococcus deserti sp. nov., a Gamma-Radiation-Tolerant Bacterium Isolated from the Sahara Desert, A. de Groot, V. Chapon, P. Servant, R. Christen, M. Fischer-Le Saux, S. Sommer, and T. Heulin, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2441-2446.
22. Gramella portivictoriae sp. nov., a Novel Member of the Family Flavobacteriaceae Isolated from Marine Sediment, S.C.K. Lau, M.M.Y. Tsoi, X. Li, I. Plakhotnikova, S. Dobretsov, P-K. Wong, and P-Y. Qian, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2497-2500.
21. Pontibacter actiniarum gen. nov., sp. nov., a Novel Member of the Phylum ‘Bacteroidetes’, and Proposal of Reichenbachiella gen. nov. as a Replacement for the Illegitimate Prokaryotic Generic name Reichenbachia Nedashkovskaya et al. 2003, O.I. Nedashkovskaya, S.B. Kim, M. Suzuki, L.S. Shevchenko, M.S. Lee, K.H. Lee, M.S. Park, G.M. Frolova, H.W. Oh, K.S. Bae, H-Y. Park, and V.V. Mikhailov, International Journal of Systematic and Evolutionary Microbiology, 2005, v.55, pp. 2583-2588.
20. Molecular Characterisation of Xanthomonas Strains Isolated from Aroids in Mauritius, M.H.R. Khoodoo, F. Sahin, M.F. Donmez, and Y. Jaufeerally Fakim, Systematic and Applied Microbiology, 2005, v.28, pp. 366-380.
19. Phylogenetic and Functional Diversity of Bacterioplankton during Alexandrium spp. Blooms, M.M. Sala, V. Balague, C. Pedras-Alio, R. Massana, J. Felipe, L. Arin, H. Illoul, and M. Estrada, FEMS Microbiology Ecology, 2005, v.54, pp. 257-67.
18. Jannaschia seosinensis sp. nov., Isolated from Hypersaline Water of a Solar Saltern in Korea, D.H. Choi, H. Yi, J. Chun, and B.C. Cho, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 45-49.
17. Culturable Phylogenetic Diversity of the Phylum ‘Bacteroidetes’ from River Epilithon and Coastal Water and Description of Novel Members of the Family Flavobacteriaceae: Epilithonimonas tenax gen. nov., sp. and Persicivirga xylanidelens gen. nov., sp. nov., L.A. O’Sullivan, J. Rinna, G. Humphreys, A.J. Weightman, and J.C. Fry, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 169-180.
16. Stenothermobacter spongiae gen. nov., sp. nov., a Novel Member of the Family Flavobacteriaceae Isolated from a Marine Sponge in the Bahamas, and Emended Description of Nonlabens tegetincola, S.C.K. Lau, M.M.Y. Tsoi, X. Li, I. Plakhotnikova, S. Dobretsov, M. Wu, P-K. Wong, J.R. Pawlik, and P-Y. Qian, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 181-185.
15. Shewanella halifaxensis sp. nov., a Novel Obligately Respiratory and Denitrifying Psychrophile, J-S. Zhao, D. Manno, C. Leggiadro, D. O’Neil, and J. Hawari, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 205-212.
14. Myceligenerans crystallogenes sp. nov., Isolated from Roman Catacombs, I. Groth, P. Schumann, B. Schutze, J.M. Gonzalez, L. Laiz, M-L. Suihko, and E. Stackebrandt, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 283-287.
13. Krokinobacter gen. nov., with Three Novel Species, in the Family Flavobacteriacea, S.T. Khan, Y. Nakagawa, and S. Harayama, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 323-328.
12. Methylobacterium adhaesivum sp. nov., a Methylotrophic Bacterium Isolated from Drinking Water, V. Gallego, M.T. Garcia, and A. Ventosa, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 339-342.
11. Pseudovibrio ascidiaceicola sp. nov., Isolated from Ascidians (Sea Squirts), Y. Fukunaga, M. Kurahashi, K. Tanaka, K. Yanagi, A. Yokota, and S. Harayama, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 343-347.
10. Bacillus macauensis sp. nov., a Long-Chain Bacterium Isolated from a Drinking Water Supply, T. Zhang, X. Fan, S. Hanada, Y. Kamagata, and H.P. Fang, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 349-353.
9. Thalassomonas loyana sp. nov., a Causative Agent of the White Plague-Like Disease of Corals on the Eilat Coral Reef, F.L. Thompson, Y. Barash, T. Sawabe, G. Sharon, J. Swings, and E. Rosenberg, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 365-368.
8. Patulibacter minatonensis gen. nov., sp. nov., a Novel Actinobacterium Isolated Using an Agar Medium Supplemented with Superoxide Dismutase, and Proposal of Patulibacteraceae fam. nov., Y. Takahashi, A. Matsumoto, K. Morisaki, and S. Omura, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 401-406.
7. Chryseobacterium soldanellicola sp. nov. and Chryseobacterium taeanense sp. nov., Isolated from Roots of Sand-Dune Plants, M.S. Park, S.R. Jung, K.H. Lee, M-S. Lee, J.O. Do, S.B. Kim, and K.S. Bae, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 433-438.
6. Acaricomes phytoseiuli gen. nov., sp. nov., Isolated from the Predatory Mite Phytoseiulus persimilis, R. Pukall, P. Schumann, C. Schütte, R. Gols, and M. Dicke, International Journal of Systematic and Evolutionary Microbiology, 2006, v.56, pp. 465-469.
5. Biolog: Modern Phenotypic Microbial Identification, B.R. Bochner, Encyclopedia of Rapid Microbiological Methods, 2006, v.2, Ch. 3, pp. 55-73.
4. Description of Xenorhabdus khoisanae sp. nov., the symbiont of the entomopathogenic nematode Steinernema khoisanae, Tiarin Ferreira, Carol A. van Reenen, Akihito Endo, Cathrin Spröer, Antoinette P. Malan and Leon M. T. Dicks, International Journal of Systematic and Evolutionary Microiology, March 2013
3. Efficacy of native antagonistic bacterial isolates in biological control of crown gall disease in Egypt, I.H. Tolba, M.A. Soliman, Annals of Agricultural Sciences, March 2013
2. Antimicrobial Resistance in Enterococcus spp. Isolated from Environmental Samples in an Area of Intensive Poultry Production, Vesna Furtula, Charlene R. Jackson, Erin Gwenn Farrell, John B. Barrett, Lari M. Hiott and Patricia A. Chambers, International Journal of Environmental Research Public Health, 2013
1. Phylogenetic signal in phenotypic traits related to carbon source assimilation and chemical sensitivity in Acinetobacter species, A. Assche, S. Álvarez-Pérez, A. Breij, J. Brabanter, K. Willems, L. Dijkshoorn, B. Lievens, Applied Microbiol Biotechnology, October 2016



