华粤企业集团有限公司
华粤行仪器有限公司
电话:020-34821111 (50条线)
传真:020-34820098 (销售部)
020-34829878 (维修部)
邮编:511442
地址:广州市番禺区兴南大道483号华粤大厦









请登录biolog官网查询:https://www.biolog.com/support/bibliography/
352. Farooq AS, Greetham D, Somani A, Marvin ME, Louis EJ, et al. (2018) Identification of Ethanologenic Yeast Strains from Wild Habitats. J Appl Microbiol Biochem. Vol.2 No.3:9
351. Viti C1, Decorosi F, Marchi E, Galardini M, Giovannetti L. High-throughput phenomics. Methods Mol Biol. 2015;1231:99-123. doi: 10.1007/978-1-4939-1720-4_7.
350. Mackie AM1, Hassan KA, Paulsen IT, Tetu SG. Biolog Phenotype Microarrays for phenotypic characterization of microbial cells. Methods Mol Biol. 2014;1096:123-30. doi: 10.1007/978-1-62703-712-9_10.
349. Chaturvedi V1,2, DeFiglio H1, Chaturvedi S1,2. Phenotype profiling of white-nose syndrome pathogen Pseudogymnoascus destructans and closely-related Pseudogymnoascus pannorum reveals metabolic differences underlying fungal lifestyles. Version 2. F1000Res. 2018 May 25 [revised 2018 Jul 17];7:665. doi: 10.12688/f1000research.15067.2. eCollection 2018.
348. The Two-Component System ZraPSR Is a Novel ESR that Contributes to Intrinsic Antibiotic Tolerance in Escherichia coli., Rome, Borde, Taher, Cayron, Lesterlin, Gueguen, De Rosny, Rodrigue, National Center for Biotechnology Information, December 2018.
347. Members of Marinobacter and Arcobacter Influence System Biogeochemistry During Early Production of Hydraulically Fractured Natural Gas Wells in the Appalachian Basin, Morgan V. Evans, Jenny Panescu, Andrea J. Hanson, Susan A. Welch, Julia M. Sheets, Nicholas Nastasi, Rebecca A. Daly, David R. Cole, Thomas H. Darrah, Michael J. Wilkins, Kelly C. Wrighton, and Paula J. Mouser, Frontiers in Microbiology, November 2018
346. Light spectrum modifies the utilization pattern of energy sources in Pseudomonas sp. DR 5-09, S. Gharaie, L. Vaas, A. Rosberg, S. Windstam, M. Karlsson, K. Bergstrand, S. Khalil, W. Wohanka, B. Alsanius, PLOS One, December 2017
345. The genome and phenome of the green alga Chloroidium sp. UTEX 3007 reveal adaptive traits for desert acclimatization, D. Nelson, B. Khraiwesh, W. Fu, S. Alseekh, A. Jaiswal, A. Chaiboonchoe, K. Hazzouri, M. O’Connor, G. Butterfoss, Genomics and evolutionary biology, plant biology, June 2017
344. Colwellia psychrerythraea Strains from Distant Deep Sea Basins Show Adaptation to Local Conditions, S. Techtmann, K. Fitzgerald, S. Stelling, D. Joyner, S. Uttukar, A. Harris, N. Alshibli, S. Brown and T. Hazen, Frontiers in Environmental Science, May 2017
343. The UbiI (VisC) aerobic ubiquinone synthase is required for expression of type 1 pili, biofilm formation, and pathogenesis in uropathogenic Escherichia coli, K. Floyd, C. Mitchell, A. Eberly, S. Colling, E. Zhang, Journal of Bacteriology, May 2017
342. Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae, C. Blin, V. Passet, M. Touchon, E. Rocha, and S. Brisse, Environmental Microbiology, 2017
341. Phenotypic Profiling Reveals that Candida albicans Opaque Cells Represent a Metabolically Specialized Cell State Compared to Default White Cells, I. Ene, M. Lohse, A. Vladu, J. Morschhäuser, A. Johnson, R. Bennett, American Society for Microbiology, Nov. 2016
340. Algal Cell Factories: Approaches, Applications, and Potentials, W. Fu, A. Chaiboonchoe, B. Khraiwesh, D. Nelson, D. Al-Khairy, A. Mystikou, A. Alzahmi and K. Salehi-Ashtiani, Marine Drugs, 2016
339. Lactobacillus oligofermentans glucose, ribose and xylose transcriptomes show higher similarity between glucose and xylose catabolism-induced responses in the early exponential growth phase, M. Andreevskaya , P. Johansson, E. Jääskeläinen, T. Rämö, J. Ritari, L. Paulin, J. Björkroth and P. Auvinen, BMC Genomics, 2016
338. Heat Survival and Phenotype Microarray Profiling of Salmonella Typhimurium Mutants, Dawoud TM, Khatiwara A, Park SH, Davis ML, Baker CA, Ricke SC, Kwon YM, Current Microbiology, Dec 2016
337. Metabolic parameters linked by phenotype microarray to acid resistance profiles of poultry-associated Salmonella enterica, J. Guard, M. Rothrock, D. Shah, D. Jones, R. Gast, R. Sanchez-Ingunza, M. Madsen, J. El-Attrache, B. Lungu, Research in Microbiology, Dec. 2016
336. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae, J Brewster, J McKellar, T Finn, J Newman, T Peat, M Gerth, Nature Oct 2016
335. Genome-Scale Metabolic Model for the Green Alga Chlorella vulgaris UTEX 395 Accurately Predicts Phenotypes under Autotrophic, Heterotrophic, and Mixotrophic Growth Conditions, C. Zuñiga, C. Li, T. Huelsman, J. Levering, D. Zielinski, B. McConnell, C. Long, E. Knoshaug, M. Guarnieri, M. Antoniewicz, M. Betenbaugh, and K. Zengler, Plant Physiology, July 2016
334. Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli, C. Engl, J. Schaefer, L. Kotta-Loizou, M. Buck, Nucleic Acids Research Advance Access, July 8, 2016
333. Pseudomonas aeruginosa MifS-MifR Two-Component System Is Specific for α-Ketoglutarate Utilization, G. Tatke, H. Kumari, E. Silva-Herzog, L. Ramirez, K. Mathee, PlosONE, June 2016
332. Phenotype microarray analysis of the drug efflux systems in Salmonella enterica serovar Typhimurium, S. Yamasaki, T. Fujioka, K. Hayashi, S. Yamasaki, M. Hayashi-Nishino, K. Nishino, Journal of Infection and Chemotherapy, 2016
331. Indispensability of Horizontally Transferred Genes and Its Impact on Bacterial Genome Streamlining, I. Karcagi, G. Draskovits, K. Umenhoffer, G. Fekete, K. Kovács, O. Méhi, G. Balikó, B. Szappanos, Z. Györfy, T. Fehér, B. Bogos, F. Blattner, C. Pál, G. Pósfai, and B. Papp, Oxford Journal, Jan 2016
330. Genomic and phenotypic characterization of the species Acinetobacter venetianus, M. Fondi, I. Maida, E. Perrin, V. Orlandini, L. La Torre, E. Bosi, Scientific Reports, Feb. 2016
329. Contribution of the Salmonella enterica KdgR Regulon to Persistence of the Pathogen in Vegetable Soft Rots, A. Georgea, I. Gonzáleza, G. Lorcab and M. Teplitskia, Applied and Environmental Microbiology, 2016
328. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating, R. García-Contreras, L. Nuñez-López, R. Jasso-Chávez, B. Kwan, J. Belmont, A. Rangel-Vega, T. Maeda and T. Wood, ISME Journal 2015,
327. Influence of quorum sensing in multiple phenotypes of the bacterial pathogen Chromobacterium violaceum, O. Mejía, C. Juárez, M. Vázquez, S. Hernandez, G. Contreras, FEMS 2014,
326. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases, R Caspi, R Billington, L Ferrer, H Foerster, C Fulcher, I Keseler, A Kothari, M Krummenacker, M Latendresse, L Mueller, Q Ong, S Paley, P Subhraveti, D Weaver and P Karp, Nucleic Acids Research, 2016,
325. Emergence of Antimicrobial-Resistant Escherichia coli of Animal Origin Spreading in Humans, D Skurnik, O Clermont, T Guillard, A Launay, O Danilchanka, S Pons, L Diancourt, F Lebreton, K Kadlec, D Roux, D Jiang, S Dion, H Aschard, M Denamur, C Cywes-Bentley, S Schwarz, O Tenaillon, A Andremont, B Picard, J Mekalanos, S Brisse and E Denamur, Molecular Biology and Evolution, Dec 2015,
324. Genome and Phenotype Microarray Analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: Genetic Determinants and Metabolic Abilities with Environmental Relevance, A Orro, M Cappelletti, P D’Ursi, L Milanesi, A Canito, J Zampolli, E Collina, F Decorosi, C Viti, S Fedi, A Presentato, D Zannoni, P Gennaro, PlosONE, Oct 2015,
323. The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self-defence, Zhang J, Bayram Akcapinar G, Atanasova L, Rahimi MJ, Przylucka A, Yang D, Kubicek CP, Zhang R, Shen Q, Druzhinina IS, Environmental Microbiology, 2015,
322. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients, Rasmus Lykke Marvig, Daniela Dolce, Lea M. Sommer, Bent Petersen, Oana Ciofu, Silvia Campana, Soren Molin, Giovanni Taccetti and Helle Krogh Johansen, BMC Microbiology, 2015,
321. Novel R Pipeline for Analyzing Biolog Phenotypic Microarray Data, M Vehkala, M Shubin, T Connor, N Thomson, J Corander, PLOSONE, 2015,
320. Multiple antibiotic susceptibility of polyphosphate kinase mutants (ppk1 and ppk2) from Pseudomonas aeruginosa PAO1 as revealed by global phenotypic analysis, J Ortiz-Severin, M Varas, C Bravo-Toncio, N Guiliani, F Chavez, Biological Research, 2015,
319. Chromosomal position shift of a regulatory gene alters the bacterial phenotype, V Gerganova, M Berger, E Zaldastanishvili, P Sobetzko, C Lafon, M Mourez, A Travers, G Muskhelishvili, Nucleic Acids Research, 2015,
318. Phenotype MicroArrays as a complementary tool to next generation sequencing for characterization of tree endophytes, K Blumenstein, D Macaya-Sanz, J Martin, B Albrectsen, JWitzell1, METHODS, 2015
317. Comparative genomic and phenomic analysis of Clostridium difficile and Clostridium sordellii, two related pathogens with differing host tissue preference. Joy Scaria, Haruo Suzuki, et al., BMC Genomics, 2015,
316. Expression of RCK2 MAPKAP (MAPK-activated protein kinase) rescues yeast cells sensitivity to osmotic stress. V Kumar, A J Hart, T T Wimalasena1, G A Tucker1 and D Greetham, Microbial Cell Factories, 2015,
315. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Qiongqiong Yan, Karen Power, et al, Frontiers in Microbiology, 2013,
314. Evaluation of butyrate-induced production of a mannose-6-phosphorylated therapeutic enzyme using parallel bioreactors. Madhavarao CN; Agarabi CD, et al, Biotechnol Appl Biochem, 2014,
313. Phenotype Microarray Analysis of Mycobacterium smegmatis and Its Isogenic Strains Reveals Novel Functions of (p)ppGpp and c-di-GMP in Mycobacterial Physiology. Kuldeepkumar Ramnaresh Gupta, Sanjay Kasetty and Dipankar Chatterji, APM, January 2015,
312. Comparative nutritional and chemical phenome of Clostridium difficile isolates determined using phenotype microarrays. Scaria J, Chen JW, et al, Int J Infect Dis, August 2014,
311. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. Hiroshi Ogasawara, Seina Ohe, Akira Ishihama, FEMS, December 2014,
310. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Kark A. Hassnan, Scott M. Jackson, et al, PNAS, September 2013,
309. Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable Pseudomonas aeruginosa in Long-term Cystic Fibrosis Infections. MSfoia Feliziani, Rasmus L. Marvig, et al, Plos One, October 2014,
308. Phenotype microarray technology and its application in industrial biotechnology, Greetham D, Biotechnol Lett, June 2014,
307. Development of a phenotypic assay for characterisation of ethanologenic yeast strain sensitivity to inhibitors released from lignocellulosic feedstocks. Greetham D, Wimalasena T, et al, J Ind Microbiol Biotechnol, June 2014,
306. Phenotypic characterisation of Saccharomyces spp. for tolerance to 1-butanol. Zaki AM, Wimalasena TT, Greetham D, Epub, September 2014,
305. High-Throughput Screening of Dipeptide Utilization Mediated by the ABC Transporter DppBCDF and Its Substrate-Binding Proteins DppA1-A5 in Pseudomonas aeruginosa. Daniel Pletzer, Corinne Lafon, et al, Plos One, October 2014,
304. L-Histidine Inhibits Biofilm Formation and FLO11-Associated Phenotypes in Saccharomyces cerevisiae Flor Yeasts . Marc Bou Zeidan, Giacomo Zara, et al, Plos One, November 2014,
303. Environmental Heterogeneity Drives Within-Host Diversification and Evolution of Pseudomonas aeruginosa. Trine Markussen, Rasmus Lykke Marvig, et al, Mol Bio No. 5, September 2014,
302. Long-term phenotypic evolution of bacteria. Germ?n Plata, Christopher S. Henry & Dennis Vitkup, Nature, September 2014,
301. Differences in carbon source utilisation distinguish Campylobacter jejuni from Campylobacter coli. Sariqa Wagley1, Jane Newcombe et al, BMC Microbiology 2014,
300. The Transcription Factor Ste12 Mediates the Regulatory Role of the Tmk1 MAP Kinase in Mycoparasitism and Vegetative Hyphal Fusion in the Filamentous Fungus Trichoderma atroviride. Sabine Gruber, Susanne Zeilinger, Plos One, Oct 2014,
299. GenoBase: comprehensive resource database of Escherichia coli K-12. Otsuka Y, Muto A, et al. Nucleic Acids Res, Nov 2014,
298. Evidence of Metabolis Switching and Implications for Food Safety from the Phenome(s) of Salmonella enterica Serovar Typhimurium DT104 Cultured at Selected Points across the Pork Production Food Chain. Marta Martins, Matthew P. McCusker, et al. Applied and Environmental Microbiology, June 2013,
297. Utilization of host-derived cysteine-containing peptides overcomes the restricted sulphur metabolism of Campylobacter jejuni. Hanne Vorwerk, Juliane Mohr, et al., Molecular Microbiology, August 2014,
296. Phenotypic Microarrays Suggest Escherichia coli ST131 Is Not a Metabolically Distinct Lineage of Extra-Intestinal Pathogenic E. coli. A Alqasim, R Emes, G Clark, J Newcombe, R La Ragione, A McNally, Plos One, Feb 2014,
295. Genotype and phenotypes of an insteine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization. Fabich AJ, Leatham MP, Grissom JE, Wiley G, Lai H, Najar F, Roe BA, Cohen PS, Conway T, Infection and Immunity, June 2011,
294. Parallel independent evolution of pathogenicity within the genus Yersinia. Sandra Reuter et al., Proceedings of the National Academy of Sciences, May 2014,
293. Rosenbergiella australoborealis sp. nov, Rosenbergiella collisarenosi sp. nov. and Rosenbergiella epipactidis sp. nov.,three novel bacterial species isolated from ?oral nectar. Marijke Lenaerts et al., Systematic and Applied Microbiology, March 2014,
292. Characterization of tetA-like gene encoding for a MFS efflux pump in Streptococcus thermophilus. S Arioli, S Guglielmetti, S Amalfitano, C Viti, E Marchi, F Decorosi, L Giovannetti and D Mora, FEMS Microbiology Letters, April 2014,
291. iOD907, the first genome-scale metabolic model for the milk yeast Kluyveromyces lactis. Oscar Dias1, Rui Pereira1, Andreas K. Gombert2, Eugenio C Ferreira and Isabel Rocha1, Biotechnology Journal, April 2014,
290. Integrative Conjugative Element ICE-Box Confers Oxidative Stress Resistance to Legionella pneumophila In Vitro and in Macrophages. Kaitlin J. Flynn, Michele S. Swanson, mBio, April 2014,
289. Genomic and Phenotypic Characterization of Vibrio cholerae Non-O1 Isolates from a US Gulf Coast Cholera Outbreak. Bradd J. Haley et al., PLOS One, April 2014,
288. Complete genome sequence of the Phaeobacter gallaeciensis type strain CIP 105210T (= DSM 26640T = BS107T).Oliver Frank1, Silke Pradella1, Manfred Rohde2, Carmen Scheuner1, Hans-Peter Klenk1, Markus Goker1, Jorn Petersen1, Standards in Genomic Sciences, March 2014,
287. Role of Intracellular Carbon Metabolism Pathways in Shigella flexneri Virulence. E. A. Waligora, C. R. Fisher, N. J. Hanovice, A. Rodou, E. E. Wyckoff and S. M. Payne, American Society for Microbiology, April 2014,
286. Phenotype microarray technology and its application in industrial biotechnology. Darren Greetham, European Union 2014, January 2014,
285. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Olga Jeske, Mareike Jogler, Jorn Petersen, Johannes Sikorski, Christian Jogler, Springer Science+Business Media Dordrecht 2013, August 2013,
284. Metabolic Model Refinement Using Phenotypic Microarray Data. Pratish Gawand, Laurence Yang, William R. Cluett, Radhakrishnan Mahadevan, Methods in Molecular Biology, May 2013,
283. Opm: An R package for analysing OmniLog Phenotype MicroArray data. Lea A. I. Vaas, Johannes Sikorski, Benjamin Hofner, Anne fiebig, Nora Buddruhs, Hans-Peter Klenk, Markus Goker, Bioinformatics Advance Access, June 2013,
282. Impacts of pr-10a Overexpression at the Molecular and the Phenotypic Level. Lea A. I. Vaas, Maja Marheine, Johannes Sikorski, Markus Goker, Heinz-Martin Schumacher, International Journal of Molecular Sciences, July 2013,
281. Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Yusuke Minato, Sara R. Fassio, Alan J. Wolfe, Claudia C. Hase, Microbiology, February 2013,
280. Phenotypic characterization of sarR mutant in Staphylococcus aureus. Andrea L. Lucas, Adhar C. Manna, Microbial Pathogenesis, November 2012,
279. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J. J. Rasmussen, C. S. Vegge, H. Frokiaer, R. M. Howlett, K. A. Krogfelt, D. J. Kelly and H. Ingmer, Journal of Medical Microbiology, May 2013,
278. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Qiongqiong Yan, Karen A. Power, Shane Cooney, Edward Fox, Gopal R. Gopinath, Christopher J. Grim, Ben D. Tall, Matthew P. McCusker, Seamus Fanning, Frontiers in Microbiology, September 2013,
277. Isolation and Characterization of Burkholderia rinojensis sp. nov., a Non-Burkholderia cepacia Complex Soil Bacterium with Insecticidal and Miticidal Activities. Ana Lucia Cordova-Kreylos, Lorena E. Fernandez, Marja Koivunen, April Yang, Lina Flor-Weiler, Pamela G. Marrone, Applied and Environmental Microbiology, October 2013,
276. Salmonella Utilizes d-Glucosaminate via a Mannose Family Phosphotransferase System Permease and Associated Enzymes. Katherine A. Miller, Robert S. Phillips, Jan Mrazek, Timothy R. Hoover, Journal of Bacteriology, July 2013,
275. Virulence and Metabolic Characteristics of Salmonella enterica Serovar Enteritidis Strains with Different SefD Variants in Hens. Cesar A. Morales, Jean Guard, Roxana Sachez-Ingunza, Devendra H. Shah, Mark Harrison, Applied and Environmental Microbiology, June 2012,
274. Dimethyl adenosine transferase (KsgA) deficiency in Salmonella entericaSerovar Enteritidis confers susceptibility to high osmolarity and virulence attenuation in chickens. Kim Lam Chiok, Tarek Addwebi, Jean Guard, Devendra H. Shah, Applied and Environmental Microbiology, October 2013,
273. Exidence of Metabolic Switching and Implications for Food Safety from the Phenome(s) of Salmonella enterica Serovar Typhimurium DT104 Cultured at Selected Points across the Pork Production Food Chain. Marta Martins, Matthew P. McCusker, Evonne M. McCabe, Denis O’Leary, Geraldine Duffy, Seamus Fanning, Applied and Environmental Microbiology, June 2013,
272. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Brooke A. Napier, Lena Meyer, James E. Bina, Mark A. Miller, Anders Sjostedt, David S. Weiss, PNAS, September 2012,
271. Genomic diversity and fitness of E. Coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Swaine L. Chen, Meng Wu, Jeffrey P. Henderson, Thomas M. Hooton, Micheal E. Hibbing, Scott J. Hultgren, Jeffrey I. Gordon, Sci Transl Med, May 2013,
270. The Transcriptional Response of Cryptococcus neoformans to Ingestion by Acanthamoeba castellanii and Macrophages Provides Insights into the Evolutionary Adaptation to the Mammalian Host. Lorena da S. Derengowski, Hugo Costa Paes, Patricia Albuquerque, Aldo Henrique F. P. Tavares, Larissa Fernandes, Ildinete Silva Pereira, Arturo Casadevall, Eukaryotic Cell 2013, March 2013,
269. Systems-Based Approaches to Probing Metabolic Variation within the Mycobacterium tuberculosis Complex. Emma K Lofthouse, Paul R Wheeler, Dany J V Beste, Bhagwati L Khatri, Huihai Wu, Ton A Mendum, Andrzej M Kierzek, Johnjoe McFadden, PLOS ONE, Volume 8, Issue 9, September 2013,
268. Β-phenylethylamine as a novel nutrient treatment to reduce bacterial contamination due to Escherichia coli O157:H7 on beef meat. Ty Lynnes, S.M. Horne, B.M. PrüΒ, Meat Science, Volume 96, Issue 1, January 2013, Pages 165-171
267. The Carbohydrate Metabolism Signature of Lactococcus lactis Strain A12 Reveals Its Sourdough Ecosystem Origin, Delphine Passerini, Michele Coddeville, Pascal Le Bourgeois, Pascal Loubiere, Paul Ritzenthaler, Catherine Fontagne-Faucher, Marie-Line Daveran-Mingot, Muriel Cocaign-Bousquet, Applied and Environmental Microbiology, Volume 79, Number 19, October 2013
266. Establishment of a Method To Rapidly Assay Bacterial Persister Metabolism, Mehmet A Orman, Mark P Brynildsen, Antimicrobial Agents and Chemotherapy, Volume 57, Number 9, September 2013, Pages 4398-4409
265. Nutritional physiology of a rock-inhabiting, model microcolonial fungus from an ancestral lineage of the Chaetothyriales (Ascomycetes), Corrado Nai, Helen Y. Wong, Annette Pannenbecker, William J. Broughton, e, Isabelle Benoit, Ronald P. de Vries, Cécile Gueidan, Anna A. Gorbushina, Fungal Genetics and Biology, Volume 56, July 2013, Pages 54-66
264. Genomic Comparison of Escherichia coli O104:H4 Isolates from 2009 and 2011 Reveals Plasmid, and Prophage Heterogeneity, Including Shiga Toxin Encoding Phage stx2, Ahmed SA, Awosika J, Baldwin C, Bishop-Lilly KA, Biswas B, et al. (2012) Genomic Comparison of Escherichia coli O104:H4 Isolates from 2009 and 2011 Reveals Plasmid, and Prophage Heterogeneity, Including Shiga Toxin Encoding Phage stx2. PLoS ONE
263. Biomimetic Synthesis of Selenium Nanospheres by Bacterial Strain JS-11 and Its Role as a Biosensor for Nanotoxicity Assessment: A Novel Se-Bioassay, Sourabh Dwivedi, Abdulaziz A. AlKhedhairy, Maqusood Ahamed, Javed Musarrat, 2013, Plos ONE
262. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia, Andrea M Sass, Crystal Schmerk, Kirsty Agnoli, Phillip J Norville, Leo Eberl, Miguel A Valvano and Eshwar Mahenthiralingam, The ISME Journal, 2013
261. The Complete Genome and Phenome of a Community-Acquired Acinetobacter baumannii, Daniel N. Farrugia, Liam D. H. Elbourne, Karl A. Hassan, Bart A. Eijkelkamp, Sasha G. Tetu, Melissa H. Brown, Bhumika S. Shah, Anton Y. Peleg, Bridget C. Mabbutt, Ian T. Paulsen, March 2013, PLoS ONE
260. Characterization of MSMEG_2631 gene (mmp) encoding a Multidrug and Toxic Compound Extrusion (MATE) family protein in Mycobacterium smegmatis, and exploration of its polyspecific nature using Biolog Phenotype MicroArray, Mukti Nath Mishra, Lacy Daniels, Department of Pharmaceutical Sciences, Texas A&M Health Science Center Irma Lerma Rangel College of Pharmacy, 1010 West Ave. B, Kingsville, Texas 78363, USA, Journal of Bacteriology 2013 Jan 4; 0
259. Pseudomonas aeruginosa PA1006, Which Plays a Role in Molybdenum Homeostasis, Is Required for Nitrate Utilization, Biofilm Formation, and Virulence, Filiatrault MJ, Tombline G, Wagner VE, Van Alst N, Rumbaugh K, et al. (2013) PLoS ONE
258. Elucidating the Regulon of the Multi Drug Resistance Regulator RarA in K. pneumoniae, Shyamasree De Majumdar1, Mark Veleba1, Sarah Finn, Seamus Fanning and Thamarai Schneiders, Antimicrob. Agents Chemother. 14 January 2013
257. Variable Carbon Catabolism among Salmonella enterica Serovar Typhi Isolates, Lay Ching Chai, Boon Hong Kong, Omar Ismail Elemfareji, Kwai Lin Thong, PlosOne May 2012, Volume 7, Issue 5
256. The Escherichia coli SLC26 homologue YchM (DauA) is a C4-dicarboxylic acid transporter, Eleni Karinou, Emma L. R. Compton, Melanie Morel, Arnaud Javelle, Molecular Microbiology Volume 87, Issue 3, pages 623 to 640, February 2013
255. Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter to quinolone susceptibility, Nadia Tocci, Francesco Iannelli, Alessandro Bidossi, Maria Laura Ciusa1, Francesca Decorosi, Carlo Viti, Gianni Pozzi1, Susanna Ricci and Marco Rinaldo Oggioni
254. The Escherichia coli SLC26 homologue YchM (DauA) is a C4-dicarboxylic acid transporter, E Karinou; ELR Compton; M Morel; A Javelle; 2012, Molecular Microbiology
253. Role of two-component sensory systems of Salmonella enterica serovar Dublin in the pathogenesis of systemic salmonellosis in cattle2, Pullinger GD; van Diemen PM; Dziva F; Stevens MP; 2012 Oct 5, p.3108-3122, Microbiology
252. Selection of bacteria capable of dissimilatory reduction of Fe(III) from a long-term continuous culture on molasses and their use in a microbial fuel cell2, Sikora A; Wojtowicz-Sienko J; Piela P; Zielenkiewicz U; Tomczyk-Zak K; Chojnacka A; Sikora R; Kowalczyk P; Grzesiuk E; Blaszczyk M; 2012 Oct, p.305-316, J Microbiol Biotechnol
251. Biolog phenotype microarrays1, Shea A; Wolcott M; Daefler S; Rozak DA; 2012 May, p.331-373, Methods Mol Biol
250. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics1, Vaas LA; Sikorski J; Michael V; Goker M; Klenk HP; 2012 Mar 6, PLoS One
249. Effects of the ERES pathogenicity region regulator Ralp3 on Streptococcus pyogenes serotype M49 virulence factor expression1, Siemens N; Fiedler T; Normann J; Klein J; Munch R; Patenge N; Kreikemeyer B; 2012 Mar, p.3618-3626, Journal of Bacteriology
248. Variable carbon catabolism among Salmonella enterica serovar Typhi isolates3, Chai LC; Kong BH; Elemfareji OI; Thong KL; 2012 Jun 19, PLoS One
247. Efficacy of biocides used in the modern food industry to control salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds2, Condell O; Iversen C; Cooney S; Power KA; Walsh C; Burgess C; Fanning S; 2012 Jun, p.3087-3097, Appl Environ Microbiol
246. Global transcriptome response to ionic liquid by a tropical rain forest soil bacterium, Enterobacter lignolyticus1, Khudyakov JI; D’haeseleer P; Borglin SE; DeAngelis KM; Woo H;Lindquist EA; Hazen TC; Simmons BA; Thelen MP; 2012 Jul, p.E2173-E2182, Proc Natl Acad Sci U S A
245. Coordinated phenotype switching with large-scale chromosome flip-flop inversion observed in bacteria3, Cui L; Neoh HM; Iwamoto A; Hiramatsu K; 2012 Jan, p.E1647-E1656, Proc Natl Acad Sci U S A
244. Phenotypic and transcriptomic response of auxotrophic Mycobacterium avium subsp. paratuberculosis leuD mutant under environmental stress9, Chen JW; Scaria J; Chang YF; 2012 Feb, p.e37884, PLoS One
243. Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments1, Chaudhry V; Rehman A; Mishra A; Chauhan PS; Nautiyal CS; 2012 Feb, p.450-460, Microbial Ecology
242. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope, Weski J; Ehrmann M; 2012 Aug 7, p.3225-3233, Journal of Bacteriology
241. A novel approach combining the Calgary Biofilm Device and Phenotype MicroArray for the characterization of the chemical sensitivity of bacterial biofilms, Santopolo L; Marchi E; Frediani L; Decorosi F; Viti C; Giovannetti L; 2012 Aug, p.1023-1032, Biofouling
240. Tools to kill: Genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina, Islam MS; Haque MS; Islam MM; Emdad EM; Halim A; Hossen QM; Hossain MZ; Ahmed B; Rahim S; Rahman MS; Alam MM; Hou S; Wan X; Saito JA; Alam M; 2012 Apr, p.493-, BMC Genomics
239. Expression of Shigella flexneri gluQ-rs gene is linked to dksA and controlled by a transcriptional terminator, Caballero VC; Toledo VP; Maturana C; Fisher CR; Payne SM; Salazar JC; 2012 Apr, p.226, BMC Microbiol
238. The evolution of metabolic networks of E. coli, Baumler DJ; Peplinski RG; Reed JL; Glasner JD; Perna NT; 2012 Apr, p.182, BMC Syst Bio
237. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli, Hadjifrangiskou M; Kostakioti M; Chen SL; Henderson JP; Greene SE; Hultgren SJ; 2011 Sep, p.1516-1529, Mol Microbiol
236. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts3, Rau MH; Hansen SK; Johansen HK; Thomsen LE; Workman CT; Nielsen KF; Jelsbak L; Hoiby N; Yang L; Molin S; 2011 Sep, p.1643-1658, Environmental Microbiology
235. Evolving promiscuously, Andersson DI; 2011 Sep, p.1199-1200, Proc Natl Acad Sci USA
234. Fungal physiology – a future perspective, Wilson RA; Talbot NJ; 2011 Oct, p.3810-3815, Microbiology
233. Carbon source utilization patterns of Bacillus simplex ecotypes do not reflect their adaptation to ecologically divergent slopes in ‘Evolution Canyon’, Israel, Sikorski J; Pukall R; Stackebrandt E; 2011 Nov 18, p.38-44, FEMS Microbiology Ecology
232. Complex phenotypic assays in high-throughput screening, Clemons PA; 2011 Nov, p.334-338, Current Opinion in Chemical Biology, Curr Opin Chem Biol
231. Application of phenotypic microarrays to environmental microbiology, Borglin S; Joyner D; DeAngelis KM; Khudyakov J; D’haeseleer P; Joachimiak MP; Hazen T; 2011 Nov, p.41-48, Current Opinion in Biotechnology
230. A Functional Genomics Approach to Establish the Complement of Carbohydrate Transporters in Streptococcus pneumoniae1, Bidossi A; Mulas L; Decorosi F; Colomba L; Ricci S; Pozzi G; Deutscher J; Viti C; Oggioni MR; 2011 May 3, p.e33320, PLoS One
229. The quorum sensing diversity within and between ecotypes of Bacillus subtilis, Stefanic P; Decorosi F; Viti C; Petito J; Cohan FM; Mandic-Mulec I; 2011 May, Environmental Microbiology
228. Assessing the Contributions of the LiaS Histidine Kinase to the Innate Resistance of Listeria monocytogenes to Nisin, Cephalosporins, and Disinfectants1, Collins B; Guinane CM; Cotter PD; Hill C; Ross RP; 2011 Mar, p.2923-2929, Appl Environ Microbiol
227. Classifying compound mechanism of action for linking whole cell phenotypes to molecular targets1, Bourne CR; Wakeham N; Bunce RA; Nammalwar B; Darrell BK; Barrow WW; 2011 Mar, p.216-223, J Mol Recognit
226. Pseudomonas putida NBRIC19 dihydrolipoamide succinyltransferase (SucB) gene controls degradation of toxic allelochemicals produced by Parthenium hysterophorus5, Mishra S; Mishra A; Chauhan PS; Mishra SK; Kumari M; Niranjan A; Nautiyal CS; 2011 Mar, p.793-808, J Appl Microbiol
225. Genotype-phenotype associations in a nonmodel prokaryote, Enstrom M; Held K; Ramage B; Brittnacher M; Gallagher L; Manoil C; 2011 Mar, MBio
224. Hydrocarbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill, Kostka JE; Prakash O; Overholt WA; Green SJ; Freyer G; Canion A; Delgardio J; Norton N; Hazen TC; Huettel M; 2011 Jun, p.7962-7974, Appl Environ Microbiol
223. Absence of Membrane Phosphatidylcholine Does Not Affect Virulence and Stress Tolerance Phenotypes in the Opportunistic Pathogen Pseudomonas aeruginosa1, Malek AA; Wargo MJ; Hogan DA; 2011 Jul, p.e30829, PLoS One
222. Loss of elongation factor P disrupts bacterial outer membrane integrity1, Zou SB; Hersch SJ; Roy H; Wiggers JB; Leung AS; Buranyi S; Xie JL; Dare K; Ibba M; Navarre WW; 2011 Jul, p.413-425, Journal of Bacteriology
221. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases3, Arendt W; Hebecker S; Jager S; Nimtz M; Moser J; 2011 Jan 25, p.1401-1416, Journal of Bacteriology
220. Genomic anatomy of Escherichia coli O157:H7 outbreaks1, Eppinger M; Mammel MK; Leclerc JE; Ravel J; Cebula TA; 2011 Jan 25, p.20142-20147, Proc Natl Acad Sci USA
219. Phenotypic changes in ciprofloxacin-resistant Staphylococcus aureus1, Mesak LR; Davies J; 2011 Jan, p.785-791, Research in Microbiology
218. Shewanella knowledgebase: integration of the experimental data and computational predictions suggests a biological role for transcription of intergenic regions1, Karpinets TV; Romine MF; Schmoyer DD; Kora GH; Syed MH; Leuze MR; Serres MH; Park BH; Samatova NF; Uberbacher EC; 2011 Dec 16, p.baq012, Database (Oxford)
217. Statistical methods for QTL mapping in cereals1, Hackett CA; 2011 Dec 13, p.585-599, Plant Molecular Biology
216. The evolution of a pleiotropic fitness tradeoff in Pseudomonas fluorescens3, MacLean RC; Bell G; Rainey PB; 2011 Dec, p.8072-8077,Proc Natl Acad Sci USA
215. Evolutionary dynamics of bacteria in a human host environment2, Yang L; Jelsbak L; Marvig RL; Damkiaer S; Workman CT; Rau MH; Hansen SK; Folkesson A; Johansen HK; Ciofu O; Hoiby N; Sommer MO; Molin S; 2011 Dec, p.7481-7486, Proc Natl Acad Sci USA
214. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis1, Oberhardt MA; Puchalka J; Martins dos Santos VA; Papin JA; 2011 Aug, p.e1001116, PLoS Comput Biol
213. Characterization of three lactic acid bacteria and their isogenic ldh deletion mutants shows optimization for YATP (cell mass produced per mole of ATP) at their physiological pHs1, Fiedler T; Bekker M; Jonsson M; Mehmeti I; Pritzschke A; Siemens N; Nes I; Hugenholtz J; Kreikemeyer B; 2011 Aug, p.612-617, Appl Environ Microbiol
212. Role of VltAB, an ABC transporter complex, in viologen tolerance in Streptococcus mutans3, Biswas S; Biswas I; 2011 Apr, p.1460-1469, Antimicrobial Agents and Chemotherapy
211. Large-scale comparative phenotypic and genomic analyses reveal ecological preferences of shewanella species and identify metabolic pathways conserved at the genus level, Rodrigues JL; Serres MH; Tiedje JM; 2011 Apr, p.5352-5360, Appl Environ Microbiol
210. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development2, Holder JW; Ulrich JC; DeBono AC; Godfrey PA; Desjardins CA; Zucker J; Zeng Q; Leach AL; Ghiviriga I; Dancel C; Abeel T; Gevers D; Kodira CD; Desany B; Affourtit JP; Birren BW; Sinskey AJ; 2010 Sep, p.e1002219, PLoS Genet
209. Membrane proteases and aminoglycoside antibiotic resistance1, Hinz A; Lee S; Jacoby K; Manoil C; 2010 Oct, p.4790-4797, Journal of Bacteriology
208. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli1, Soo VW; Hanson-Manful P; Patrick WM; 2010 Nov, p.1484-1489, Proc Natl Acad Sci USA
207. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL12282, Liao YC; Huang TW; Chen FC; Charusanti P; Hong JS; Chang HY; Tsai SF; Palsson BO; Hsiung CA; 2010 May, p.1710-1717, Journal of Bacteriology
206. Differential gene expression by RamA in ciprofloxacin-resistant Salmonella Typhimurium4, Zheng J; Tian F; Cui S; Song J; Zhao S; Brown EW; Meng J; 2010 May, p.e22161, PLoS One
205. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis1, Desjardins CA; Champion MD; Holder JW; Muszewska A; Goldberg J; Bailao AM; Brigido MM; Ferreira ME; Garcia AM; Grynberg M; Gujja S; Heiman DI; Henn MR; Kodira CD; Leon-Narvaez H; Longo LV; Ma LJ; Malavazi I; Matsuo AL; Morais FV; Pereira M; Rodriguez-Brito S; Sakthikumar S; Salem-Izacc SM; Sykes SM; Teixeira MM; Vallejo MC; Walter ME; Yandava C; Young S; Zeng Q; Zucker J; Felipe MS; Goldman GH; Haas BJ; McEwen JG; Nino-Vega G; Puccia R; San-Blas G; Soares CM; Birren BW; Cuomo CA; 2010 Mar, p.e1002345, PLoS Genet
204. Cryptic prophages help bacteria cope with adverse environments1, Wang X; Kim Y; Ma Q; Hong SH; Pokusaeva K; Sturino JM; Wood TK; 2010 Mar, p.147, Nat Commun
203. Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir1, Eppinger M; Mammel MK; Leclerc JE; Ravel J; Cebula TA; 2010 Jun, p.2916-2925, Appl Environ Microbiol
202. Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile1, Franz E; van Hoek AH; Bouw E; Aarts HJ; 2010 Jun, p.8088-8096, Appl Environ Microbiol
201. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli1, Leblanc SK; Oates CW; Raivio TL; 2010 Jun, p.3367-3375, Journal of Bacteriology
200. A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes1, Damon C; Vallon L; Zimmermann S; Haider MZ; Galeote V; Dequin S; Luis P; Fraissinet-Tachet L; Marmeisse R; 2010 Jun, p.1871-1880, ISME J
199. Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni, Muraoka WT; Zhang Q; 2010 Jul 30, p.1065-1075, Journal of Bacteriology
198. Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1, Smits TH; Rezzonico F; Kamber T; Blom J; Goesmann A; Ishimaru CA; Frey JE; Stockwell VO; Duffy B; 2010 Feb, p.e22247, PLoS One
197. H2S: a universal defense against antibiotics in bacteria, Shatalin K; Shatalina E; Mironov A; Nudler E; 2010 Feb, p.986-990, Science
196. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042, Chaudhuri RR; Sebaihia M; Hobman JL; Webber MA; Leyton DL; Goldberg MD; Cunningham AF; Scott-Tucker A; Ferguson PR; Thomas CM; Frankel G; Tang CM; Dudley EG; Roberts IS; Rasko DA; Pallen MJ; Parkhill J; Nataro JP; Thomson NR; Henderson IR; 2010 Dec, p.e8801, PLoS One
195. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen, Healy B; Cooney S; O’Brien S; Iversen C; Whyte P; Nally J; Callanan JJ; Fanning S; 2010 Aug 1, p.339-350, Foodborne Pathog Dis
194. Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress, Johler S; Stephan R; Hartmann I; Kuehner KA; Lehner A; 2010 Apr, p.1053-1061, Appl Environ Microbiol
193. Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter to quinolone susceptibility, Nadia Tocci, Francesco Iannelli, Alessandro Bidossi, Maria Laura Ciusa, Francesca Decorosi, Carlo Viti, Gianni Pozzi, Susanna Ricci and Marco Rinaldo Oggioni, Published ahead of print 31 October 2012, doi: 10.1128/AAC.01298-12.
192. Global Phenotypic Characterization of Bacteria, B.R. Bochner, FEMS Microbiol. Rev., 2009, v. 33, pp.191-205.
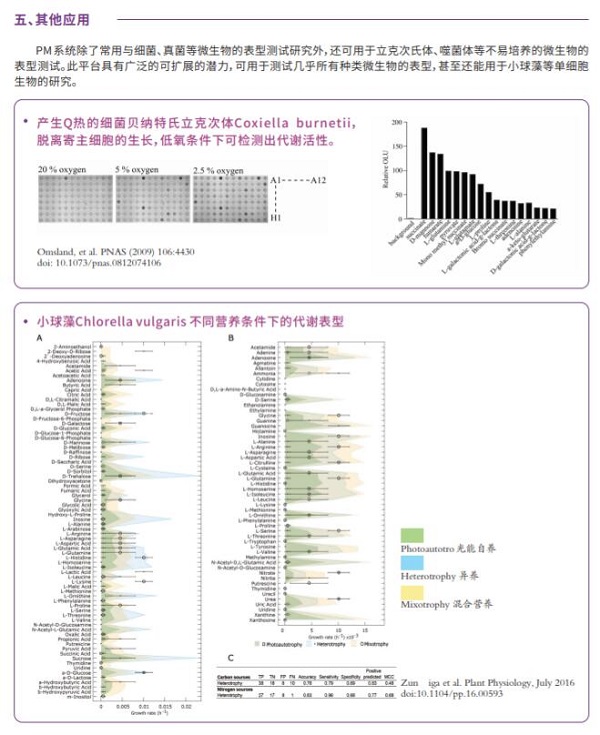
191. Host Cell-free Growth of the Q Fever Bacterium Coxiella burnetii, A. Omsland, D.C. Cockrell, D. Howe, E.R. Fischer, K. Virtaneva, D.E. Sturdevant, S.F. Porcella, R.A. Heinzen, Proceedings of the National Academy of Sciences, 2009, Epub Feb. 25.
190. Legionella pneumophila Couples Fatty Acid Flux to Microbial Differention And Virulence, R.L. Edwards, Z.D. Dalebroux, M.S. Swanson, Mol. Microbiol., 2009, v. 71, pp. 1190-1204.
189. SpoT Governs Legionella pneumophila Differentiation in Host Macrophages, Z.D. Dalebroux, R.L. Edwards, M.S. Swanson, Mol. Microbiol., 2009, v.71, pp. 640-658.
188. Involvement of the oscA Gene in the Sulphur Starvation Response and in Cr(VI) Resistance in Pseudomonas corrugata 28, C. Viti, F. Decorosi, A. Mini, E. Tatti, L. Giovannetti, Microbiology, 2009, v. 155, pp. 95-105.
187. Overcoming the Anaerobic Hurdle in Phenotypic Microarrays: Generation and Visualization of Growth Curve Data for Desulfovibrio vulgaris Hildenborough, S. Borglin, D. Joyner, J. Jacobsen, A. Mukhopadhyay, T. C. Hazen, J. Microbiol. Methods, 2008. v. 76 pp. 159-168.
186. Characterization of the myo-Inositol Utilization Island of Salmonella enterica serovar Typhimurium, C. Kroger, T.M. Fuchs, J. Bacteriol. 2009, v. 191, pp. 545-554.
185. Phenotypic Diversity among Cronobacter spp., B. Tall, 1st International Meeting on Cronobacter, 2009, Dublin, Ireland.
184. What is (and isn’t) Cronobacter, C. Iversen, 1st International Meeting on Cronobacter, 2009, Dublin, Ireland.
183. Genome-Scale Reconstruction and Analysis of the Pseudomonas putida KT2440 Metabolic Network Facilitates Applications in Biotechnology, J. Puchalka, M. A. Oberhardt, M. Godinho, A. Bielecka, D. Regenhardt, K. N. Timmis, J. A. Papin, V. A. P. Martins dos Santos, PLoS Computational Biology, 2008, v. 4.pp. 1-18.
182. High Throughput Phenotypic Characterization of Pseudomonas aeruginosa Membrane Transport Genes, D.A. Johnson, S.G. Tetu, K. Phillippy, J. Chen, Q. Ren & I.T. Paulsen, PLoS Genet., 2008, v. 4, pp. 1-11.
181. RamA Confers Multidrug Resistance in Salmonella enterica via Increased Expression of acrB, which is Inhibited by Chlorpromazine, A.M. Bailey, I.T. Paulsen, L.J. Piddock, Antimicrob Agents, 2008 v. 52, pp. 3604-3611.
180. Phenotype Profiling of Single Gene Deletion Mutants of E. coli Using Biolog Technology, Y. Tohsato, H. Mori, Proceeding of the 19th International Conference on Genome Informatics (GIW 2008), Gold Coast, Australia, pp. 1-11.
179. The Applications of Systematic In-Frame, Single-Gene Knockout Mutant Collection of Escherichia coli K-12, T. Baba, HC Huan, K. Datsenko, BL Wanner, H. Mori, Methods Mol. Biol., 2008, v. 416, pp. 183-194.
178. Identification and Characterization of Brewers Yeast Using Phenotype MicroArrays, S. Walker, Abstract, Veterinary Laboratories Agency Conference, November 2008.
177. Development of Phenotype MicroArrays for The Characterisation of a Porcine Cell Line, M. Abu Oun, Abstract, Veterinary Laboratories Agency Conference, November 2008.
176. The Gut Microbiota as a Virtual Organ And Novel Approaches to Understand it, J. Marchesi, Abstract, Veterinary Laboratories Agency Conference, November 2008.
175. Effects of Microbial Growth Rate on Inhibitory Effects of Antimicrobial Compounds Using Biolog Microarray Chemical Sensitivity Plates, R. Thorn, Abstract, Veterinary Laboratories Agency Conference, November 2008.
174. Use of Biolog for Studying Oral Communities, J. Greenman, Abstract, Veterinary Laboratories Agency Conference, November 2008.
173. Integrating PM Technology With In Situ and In Silico Analyses of Microbial Activities, G. Preston, Abstract, Veterinary Laboratories Agency Conference, November 2008.
172. Metabolomics of Slow-Growing Mycobacterium tuberculosis and Mycobacterium bovis Strains Using Phenotype MicroArray Technology, B. Upadhyay, Abstract, Veterinary Laboratories Agency Conference, November 2008.
171. Using Phenotypic Data to Construct and Curate Genome-scale Models, P.Suthers, Abstract, Veterinary Laboratories Agency Conference, November 2008.
170. The Detection of Differences in the Utilisation of Glyoxylate and 2-Carbon Compounds in Salmonella typhimurium, M. Saunders, Abstract, Veterinary Laboratories Agency Conference, November 2008.
169. Fitness Costs and Stability of a High-level Ciprofloxacin Resistance Phenotype in Salmonella enteritidis, S. Fanning, Abstract, Veterinary Laboratories Agency Conference, November 2008.
168. Phenotypic and Genotypic Differences Between Pathogenic and Laboratory Escherichia coliStrains, J. Hobman, Abstract, Veterinary Laboratories Agency Conference, November 2008.
167. Identification of an ABC-Type Trehalose Transporter in Mycobacterium smegmatis, S. Jaques III, L. Daniels, poster, SC-ASM Conference, Nov. 2008, Austin.
166. Carbon Source Utilization Pattern Among Rapidly Growing Mycobacterium (RGM) Species by Phenotypic Array Analysis Using Biolog OmniLog System, M. Rahman, S. Jaques III, L. Daniels, poster, SC-ASM Conference, Nov. 2008, Austin
165. Development of a New Test Panel for Identification of Both Gram-Negative and Gram-Positive Bacteria, A. Franco-Buff, V. Gomez, E. Olender, G. Gadzinski, B. Bochner, poster, ECCMID, April, 2008, Barcelona
164. Phenotype MicroArray Analysis of The Metabolism of Helicobacter pylori, X-H Lei, B. Bochner, poster, Spring Meeting, Society for General Microbiology, March, 2008, Edinburgh
163. Important Discoveries from Analysing Bacterial Phenotypes, B. R. Bochner, L. Giovannetti, C. Viti, Mol. Microbiol., 2008, v. 70, pp. 274-280.
162. Florence Conference on Phenotype MicroArray Analysis of Microorganisms, The Environment, Agriculture, and Human Health, C. Viti, B. Bochner, L. Giovannetti, 2008, Annals of Microbiology, v. 2, pp. 347-349.
161. Phenotypes of Salmonella enterica Serovar Typhi and Acinetobacter Spp., Reflections of Different Lifestyles?, L. Dijkshoorn, J.T. Van Dissel, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
160. Longitudinal Study of Pseudomonas aeruginosa Isolates from Cystic Fibrosis Lungs, L. Yang, S.K. Hansen, L. Jelsbak, H. Jarmer , J.A.J. Haagensen, H.K. Johansen, N. Hoiby, S. Molin, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
159. Functional Diversity and Productivity Peak at Intermediate Dispersal Rate in Evolving Bacterial Metacommunities, P. Venail, R.C. Maclean, T. Bouvier, M.A. Brockhurst, M.E. Hochberg, N. Mouquet, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
158. Adapting Biolog Phenotype MicroArray Technology to Reveal the Metabolomics of Mycobacterium tubercolosis and Mycobacterium bovis, Slow-growing Pathogens with d=0.03 to 0.05/h., B. Upadhyay, M.D. Fielder, P.R. Wheeler, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
157. Phenotypic Microarray analysis of Cronobacter Spp. (Formerly Enterobacter sakazakii), B.D. Tall, S. Fanning, C. Iversen, N. Mullane, M. H. Kothary, A. Datta, L. Carter, S. K. Curtis, B.A. McCardell, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
156. Transcriptional Response of Desulfovibrio vulgaris Hildenborough to Oxygenation: An Indirect Metabolic Analysis by Restriction Fragment Functional Display (RFFD), M. Santana, J. Gonzalez, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
155. Influence of Pseudomonas aeruginosa on Biofilm Formation and Internalization of Burkholderia cenocepacia Strains, L. Pirone, C. Auriche, C. Dalmastri, F. Ascenzioni,, A. Bevivino , Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
154. The Metabolic Fingerprint of Filamentous Fungi Responsible for Damage and Biodegradation in Cultural Heritage: Applications and Perspectives, F.P. Pinzari, V. C. Cialei, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
153. Phenotypic Characterization of Thermophilic Bacilli Isolated from the Sites with Geothermal and Geo-chemical Anomalies of Armenia, H. Panosyan, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
152. Regulatory Network of AcrAB Multidrug Efflux Pump in Salmonella and its Role in Response to Metabolites, E. Nikaido, K. Nishino, A. Yamaguchi, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
151. Roles of Multidrug Efflux Pumps in Antimicrobial Peptide Resistance of Salmonella enterica, T. Nakano, K. Nishino, A. Yamaguchi, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
150. Phenotypic Analysis of Multidrug Efflux Pumps – Not Just for Multidrug Resistance, K. Nishino, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
149. Assessment of Phenotypic Features of Bacteria Isolated from Different Antarctic Environments, S. Mangano, C. Caruso, L. Michaud, A. Lo Giudice, V. Bruni, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
148. Genotypic and Phenotypic Diversity of Streptococcus thermophilus Strains Isolated from the Caucasian Yogurt-like Product Matsoni, I. Malkhazova, M. Merabishvili, N. Chanishvili, L. Brusetti, D. Daffonchio, D. Mora, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
147. Differentiation of Novel Species Using the Phenotype MicroArray, C. Iversen, N. Mullane, S. Fanning, B. Mccardell, B.D. Tall, M.H. Kothary, L. Carter, A. Lehner, R. Stephan, H. Joosten, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
146. Phenotypic Analysis of Enterobacter sakazakii (Cronobacter spp.), C. Iversen, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
145. Use of a Phenotype Array to Study an Adaptive Response in Enterobacter sakazakii, B. Arku, S. Fanning, T. Quinn, K. Jordan, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
144. Phenotype MicroArray Analysis of Anaerobes, T. C. Hazen, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
143. A Phenotypic Approach to Differentiate Strains within Oenococcus oeni species, S. Guerrini, S. Mangani, L. Granchi, M. Vincenzini, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
142. H-NS, A Master Controller of Warm Adaptation in Escherichia coli: A Transcriptomic Analysis, F. Fabretti, T. Carzaniga, F. Falciani, G. Deho, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
141. Novel Cues of L. pneumophila Differentiation Uncovered by Phenotype MicroArrays, R.L. Edwards, Z.D. Dalebroux, M. S. Swanson, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
140. Comparative Phenotypic Microarray Analysis of Listeria monocytogenes Strains Involved in Invasive and Gastroenteritis Listeriosis Outbreaks, A. Datta, L.Carter, L. Burall, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
139. Carbon and Energy Metabolism of Escherichia coli in the Intestine, T. Conway, P.S. Cohen, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
138. Biolog Microbial Identification System-study on the Operating Regulation of Bacteria Identification, C. Chi, Y. Mei, L. Jin-Xia, Y. Su, H. Hai-Rong, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
137. Identification of Saccharomyces cerevisiae by the Biolog automated microbes identification system, C. Chi, L. Jin-Xia, Y. Su, H. Hai-Rong, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
136. Extended-spectrum Beta-lactamase Producing Strains of Escherichia coli Isolates: Phenotypic and Genotypic Characterization, E.C. Adibe, C.E. Ilodigwe, H.I. Ekenedo, L.C. Ofora, S.I. Okonkwo, S.A. Obi, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
135. Characterization of a Porcine Cell Line by Phenotype MicroArray, M. Abuoun, J.W. Collins, M.J. Woodward, R.M. La Ragione, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
134. Integrating Biolog Phenomic Analysis with Genomic Approaches to Explore the Diversity of Natural Sinorhizobium meliloti Strains, E. Biondi , Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
133. Phenotypic Analysis of Cr(VI)-sensitive Mutants of Pseudomonas corrugata Strain 28, E. Tatti, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
132. Physiological Characterization of Pseudomonas pseudoalcaligenes KF707: Biofilm Development and Adaptation to Environmental Stress, S. Fedi, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
131. Antimicrobials inducing E. coli Biofilm Formation, Identified by Phenotype MicroArrays, A. Boehm, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
130. Linking Phenotype to Genotype of Epidemiologically Prevalent Salmonella by Comparative Microarray Analysis, J. Guard-Bouldin, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
129. Phenotypic Analysis of Salmonella, M. Anjum, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
128. Use of PM Technology to Characterize Nutrient Utilization and Restriction in the Obligate Intracellular Bacterial Pathogen Coxiella burnetii, A. Omsland, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
127. Characterization of Porcine Cell Using Phenotype MicroArray, M. Abuoun, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
126. PM Assay of Mammalian Cells, B. Bochner, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
125. Phenotype MicroArrays as a Tool to Study Physiology of Industrially Important Fungi, I. Druzhinina, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
124. Pleiotropic Effects of Mutations of the LEU4 Gene in Saccharomyces cerevisiae, E. Casalone, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
123. Unravelling Membrane Transport Mechanisms in Pseudomonas sp. through Phenomics, I. Paulsen, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
122. Towards Elucidation of Physiological Networks in a Cell Using PM Technology and Genetic Network Analysis, H. Mori, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
121. Use of Growth Profiling Data to Improve Constraint-based Modelling of Metabolism, L. Yang, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008
120. Phenotype MicroArray Technology: Principles and Practice, B. Bochner, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
119. Phenotypic Analysis of Food-borne Pathogens, T. Cebula, Abstract, Florence Conference on Phenotype MicroArray Analysis of Microorganisms, March, 2008.
118. Visualization of Growth Curve Data from Phenotype Microarray Experiments,, J.S. Jacobsen, D.C. Joyner, S.E. Borglin, T.C. Hazen, A.P. Arkin, E.W. Bethel, 11th International Conference on Information Visualization (IV07), Zurich, Switzerland, July 4-6, 2007, Published by the IEEE Computer Society.
117. Response of Desulfovibrio vulgaris to alkaline stress, S. Stolyar, Q. He, M.P. Joachimiak, Z. He, Z.K. Yang, S.E. Borglin, D.C. Joyner, K. Huang, E. Alm, T.C. Hazen, J. Zhou, J.D. Wall, A.P. Arkin, D.A. Stahl, 2007, J. Bacteriol., v.189, pp. 8944-8952.
116. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough, K.S. Bender, H-C Yen, C.L. Hemme, Z. Yang, Z. He, Q. He, J. Zhou, K.H. Huang, E.J. Alm, T.C. Hazen, A.P. Arkin, J.D. Wall, 2007, Appl. Environ. Microbiol., v.73, pp. 5389-5400.
115. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe (III)-NTA, Y.J. Tang, R. Chakraborty, H. Garcia Martin, J. Chu, T.C. Hazen, J.D. Keasling, 2007, Appl. Environ. Microbiol., v.73, pp. 3859-3864.
114. Cell wide response to low oxygen exposure in Desulfovibrio vulgaris Hildenborough, A. Mukhopadhyay, A.M. Redding, M.P. Joachimiak, A.P. Arkin, S.E. Borglin, P.S. Dehal, R. Chakraborty, J.T. Geller, T.C. Hazen, D.C. Joyner, V.J.J. Martin, J.D. Wall, Z.K. Yank, J. Zhou, J.D. Keasling, J. Bacteriol., v.189, pp. 5996-6010.
113. Pathway confirmation and flux analysis using 13C isotopic labeling of metabolites in Desulfovibrio vulgaris Hildenborough via FT-ICR Mass Spectrometry, Y. Tang, F. Pingitore, A. Mukhopadhyay, R. Phan, T.C. Hazen, J.D. Keasling, 2007, J. Bacteriol., v.189, pp. 940-949.
112. Study of Nitrate Stress in Desulfovibrio vulgaris Hildenborough Using iTRAQ Proteomics, A.M. Redding, A. Mukhopadhyay, D. Joyner, T.C. Hazen, J.D. Keasling, 2006, Briefings in Functional Genomics and Proteomics, v.5, pp. 133-143.
111. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough inferred from global analysis, Q. He, K.H. Huang, Z. He, E.J. Alm, M.W. Fields, T.C. Hazen, A.P. Arkin, J.D. Wall, J. Zhou, 2006, Appl. Environ. Microbiol, v.72, pp. 4370-4381.
110. Salt stress in Desulfovibrio vulgaris Hildenborough: An integrated genomics approach, A. Mukhopadhyay, Z. He, E.J. Alm, A.P. Arkin, E.E. Baidoo, S.C. Borglin, W. Chen, T.C. Hazen, Q. He, H.-Y. Holman, K. Huang, R. Huang, D.C. Joyner, N. Katz, M. Keller, P. Oeller, A. Redding, J. Sun, Z. Yang, J.D. Wall, J. Wei, H.-C. Yen, J. Zhou, J.D. Keasling, J. Bacteriol, v.188, pp. 4068-4078.
109. The global ppGpp-mediated stringent response to amino acid starvation in Escherichia coli, M.F. Traxler, S.M. Summers, H-T Nguyen, V. M. Zacharia, G.A. Hightower, J.T. Smith, T. Conway, Molecular Microbiology, v.68, pp. 1128-1148.
108. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa, M.A. Oberhardt, J. Puchalka, K.E. Fryer, V.A. Martins dos Santos, J.A. Papin, Bacteriol., v.190, pp. 2790-2803.
107. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine, A.J. Fabich, S.A. Jones, F.Z. Chowdhury, et al., Infect. Immun., v.76, pp. 1143-1152.
106. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities, P.A. Venail, R.C. MacLean, T. Bouvier, M.A. Brockhurst, M.E. Hochberg, N. Mouquet, Nature, v.452, pp. 210-214.
105. Photostimulation of Hypocrea atroviridis growth occurs due to a cross-talk of carbon metabolism, blue light receptors and response to oxidative stress, M.A. Friedl, M. Schmoll, C.P. Kubicek, I.S. Druzhinina, Microbiology, 2008, v.154, pp. 1229-1241.
104. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis, M.A. Friedl, C.P. Kubicek, I.S. Druzhinina, Appl. Environ. Microbial., 2008, v.74, pp. 245-250.
103. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information., A.M. Feist, C.S. Henry, J.L. Reed, M. Krummenacker, A.R. Joyce, P.D. Karp, L.J. Broadbelt, V. Hatzimanikatis, B.O. Palsson, Mol. Syst. Biol., 2007, v.3, p. 121.
102. Beta-D-Allose inhibits fruiting body formation and sporulation in Myxococcus xanthus, M. Chavira, N. Cao, K. Le, T. Riar, N. Moradshahi, M. McBride, R. Lux, W. Shi, J. Bacteriol., 2007, v.189, pp. 169-178.
101. Application of DNA bar codes for screening of industrially important fungi: the haplotype of Trichoderma harzianum sensu stricto indicates superior chitinase formation., V. Nagy, V. Seidl, G. Szakacs, M. Komon-Zelazowska, C.P. Kubicek, I.S. Druzhinina, Appl. Environ. Microbiol., 2007, v.73, pp. 7048-7058.
100. Exploring Genotypic and Phenotypic Diversity of Microbes Using Microarray Approaches, A. Mukherjee, S.A. Jackson, J.E. LeClerc, T.A. Cebula, Toxicology Mechanisms and Methods, 2006, v.16, pp. 121-128.
99. Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina, I.S. Druzhinina, M. Schmoll, B. Seiboth, C.P. Kubicek, Appl. Environ. Microbiol., 2006, v.72, pp. 2126-2133.
98. Carbon source utilization by the marine Dendryphiella species D. arenaria and D. salina, T.E. dela Cruz, B.E. Schulz, C.P. Kubicek, I.S. Druzhinina, FEMS Microbial Ecol., 2006, v.58, pp. 343-353.
97. H-NS controls metabolism and stress tolerance in Escherichia coli O157:H7 that influence mouse passage, I. Erol, K-C Jeong, D.J. Baumler, B. Vykhodets, S-H Choi, C.W. Kaspar , BMC Microbiology, 2006, v.6.
96. A Screening system for carbon sources enhancing beta-N-acetylglucosaminidase formation in Hypocrea atroviridis (Trichoderma atroviride), V. Seidl, IS Druzhinina, C.P. Kubiceka, Microbiology, 2006, v.152, pp. 2003-2012.
95. Altered Utilization of N-Acetyl-D-Galactosamine by Escherichia coli O157:H7 from the 2006 Spinach Outbreak, A. Mukherjee, M. K. Mammel, J. E. LeClerc, T. A. Cebula, Journal of Bacteriology, 2008, v.190, pp. 1710-1717.
94. Uncovering New Metabolic Capabilities of Bacillus subtilis Using Phenotype Profiling of Rifampin-Resistant rpoB Mutants, A. E. Perkins, W. L. Nicholson, Journal of Bacteriology, 2008, v.190, pp. 807-814.
93. Carbon Source Dependence and Photostimulation of Conidiation in Hypocrea atroviridis, M. A. Friedl, C. P. Kubicek, I. S. Druzhinina, Applied and Environmental Microbiology, 2008, v.74, pp. 245-250.
92. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1, M. A. Oberhardt, J. Puchalka, K. E. Fryer, V. A. P. Martins dos Santos, J. A. Papin, Journal of Bacteriology, v.190, pp. 2790-2803.
91. Dual Involvement of CbrAB and NtrBC in the Regulation of Histidine Utlization in Pseudomonas fluorescens SBW25, X.-X. Zhang, P. B. Rainey, Genetics, 2008, v.190, pp. 185-195.
90. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information, A. M. Feist, C. S. Henry, J. L. Reed, M. Krummenacker, A. R. Joyce, P. D. Karp, L. J. Broadbelt, V. Hatzimanikatis, B. Paisson, Molecular Systems Biology 3, 2007.
89. Regulatory Overlap and Functional Redundancy among Bacillus subtilis Extracytoplasmic Function s Factors, T. Mascher, A.-B. Hachmann, J. D. Helmann, Journal of Bacteriology, 2007, v.189, pp. 7500-7511.
88. Pseudomonas syringae pv. tomato DC3000 Uses Constitutive and Apoplast-Induced Nutrient Assimilation Pathways to Catabolize Nutrients That Are Abundant in the Tomato Apoplast, A. Rico, G. M. Preston, MPMI, 2008, v.21, pp. 269-282.
87. Genome-scale Reconstruction of Metabolic Network in Bacillus subtilis Based on High-throughput Phenotyping and Gene Essentiality Data, Y. K. Oh, B. O. Palsson, S. M. Park, C. H. Schilling, R. Mahadevan, J. Biol. Chem., v.282, pp. 28791-28799.
86. Genomic and Phenotypic Diversity of Coastal Vibrio cholerae Strains Is Linked to Environmental Factors, D. P. Keymer, M. C. Miller, G. K. Schoolnik, A. B. Boehm, Applied and Environmental Microbiology, 2007, v.73, pp. 3705-3714.
85. Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics, M. Mols, M. de Been, M. H. Zwietering, R. Moezelaar, T. Abee, Environmental Microbiology, 2007, pp. 2933-2944.
84. Integrated bioinformatic and phenotypic analysis of RpoN-dependent traits in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25, J. Jones, D.J. Studholme, C.G. Knight, G.M. Preston, Environmental Microbiology, 2007, pp. 3046-3064.
83. Genetic Analysis of a Novel Pathway for D-Xylose Metabolism in Caulobacter crescentus, C. Stephens, B. Christen, T. Fuchs, V. Sundaram, K. Watanabe, U. Jenal, Journal of Bacteriology, 2007, v.189, pp. 2181-2185.
82. Visualization of Growth Curve Data from Phenotype Microarray Experiments, J. S. Jacobsen, D. C. Joyner, S. E. Borglin, T. C. Hazen, A. P. Arkin, E. W. Bethel, 11th International Conference Information Visualization (IV 07), 2007.
81. Contribution of Target Gene Mutations and Efflux to Decreased Susceptibility of Salmonella enterica Serovar Typhimurium to Fluoroquinolones and Other Antimicrobials, S. Chen, S. Cui, P. F. McDermott, S. Zhao, D. G. White, I. Paulsen, J. Meng, Antimicrobial Agents and Chemotherapy, 2007, v.51,pp. 535-542.
80. Cryptococcus neoformans a Strains Preferentially Disseminate to the Central Nervous System during Coinfection, K. Nielsen, G. M. Cox, A. P. Litvintseva, E. Mylonakis, S. Malliaris, D. K. Benjamin, Jr., S. S. Giles, T. G. Mitchell, A. Casadevall, J. R. Perfect, J. Heitman, Infection and Immunity, 2005, v.73, pp. 4922-4933.
79. Regulation of Carbon and Nitrogen Utilization by CbrAB and NtrBC Two-Component Systems in Pseudomonas aeruginosa, W. Li, C-D. Lu, Journal of Bacteriology, 2007, v.189, pp. 5413-5420.
78. Diversity of soil mycobacterium isolates from three sites that degrade polycyclic aromatic hydrocarbons, C.D. Miller, R. Child, J.E. Hughes, M. Benscai, P.P. Der, R.C. Sims, A.J. Anderson, Journal for Applied Microbiology, v.102, pp. 1612-1624.
77. Characterization of Chromate-Resistant and Reducing Bacteria by Traditional Means and by a High-Throughput Phenomic Technique for Bioremediation Purposes, C. Viti, F. Decorosi, E. Tatti, L. Giovannetti, Biotechnol Prog., 2007, v.23, pp. 553-559.
76. Growth Characteristics of Microorganisms on Commercially Available Animal-free Alternatives to Tryptic Soy Medium, D. Cleland, K. Jastrzembski, E. Stamenova, J. Benson, C. Catranis, D. Emerson, B. Beck, Journal of Microbiological Methods, 2007, v.69, pp.345-352.
75. Escherichia coli enzyme IIANtr Regulates the K+ Transporter TrkA, C-R. Lee, S-H. Cho, M-J. Yoon,A. Peterkofsky, Y-J. Seok, 2007, The National Academy of Sciences of the USA, 2007.
74. A Previously Undescribed Pathway for Pyrimidine Catabolism, K.D. Loh, P. Gyaneshwar, E.M. Papadimitriou, R. Fong, K-S. Kim, R. Parales, Z. Zhou, W. Inwood, and S. Kustu, Proceedings of the National Academy of Sciences, 2006, v.103, pp. 5114-5119.
73. FhuA and HgbA, Outer Membrane Proteins of Actinobacillus pleuropneumoniae: Their Role as Virulence Determinants, L. Shakarji, L.G. Mikael, R. Srikumar, M. Kobisch, J.W. Coulton, and M. Jacques, Canadian Journal of Microbiology, 2006, v.52, pp. 391-396.
72. Phenotype MicroArray Analysis of Yeast Strains, B. Bochner, Abstract from the International Wine Microbiology Symposium, 2006.
71. Phenotype MicroArrays that Assay the Energy Producing Pathways Active in Cells, B. Bochner, Abstract from the IBC Conference on Metabolic Syndrome, 2006.
70. Global Carbon Utilization Profiles of Wild-Type, Mutant, and Transformant Strains of Hypocrea jecorina, I.S. Druzhinina, M. Schmoll, B. Seiboth, and C.P. Kubicek, Applied and Environmental Microbiology, 2006, v.72, pp. 2126-2133.
69. Yeast as a Drug Discovery Platform in Huntington’s and Parkinson’s Diseases, T.F. Outeiro and F. Giorgini, Biotechnology Journal, 2006, v.1, pp. 1-12.
68. Cell Scenario a New Look at Microarrays, C. Potera, Environmental Health Perspectives, 2006, v.114, pp. A-172-A175.
67. Biolog Advances Global Cell Analysis Scans, C. Potera, Genetic Engineering News, 2006, v.26, pp. 24-25.
66. Development of Phenotype MicroArrays for Measuring Energy Producing Pathways in Mammalian Cells, L.A. Wiater, M. Siri, R. Huang, and B.R. Bochner, Poster presented at Cambridge Healthtech Institute’s 13th International Molecular Medicine Tri-Conference, February 2006 and The Federation of American Societies for Experimental Biology, April 2006.
65. Correlation of Phenotype with the Genotype of Egg-Contaminating Salmonella enterica Serovar Enteritidis, C.A. Morales, S. Porwollik, J.G. Frye, H. Kinde, and M. McClelland, Applied and Environmental Microbiology, 2005, v.71, pp. 4388-4399.
64. A selC-Associated Genomic Island of the Extraintestinal Avian Pathogenic Escherichia coli Strain BEN2908 is Involved in Carbohydrate Uptake and Virulence, I. Chouikha, P. Germon, A. Bree, P. Gilot, M. Moulin-Schouleur, and C. Schouler, Journal of Bacteriology, 2006, v.188, pp. 977-987.
63. Phenotype Microarray Profiling of Staphylococcus aureus menD and hemB Mutants with the Small-Colony-Variant Phenotype, C. von Eiff, P. McNamara, K. Becker, D. Bates, X-H. Lei, M. Ziman, B.R. Bochner, G. Peters, and R.A. Proctor, Journal of Bacteriology, 2006, v.188, pp. 687-693.
62. Discovering New Drugs on the Cellular Level, NASA Spinoff, 2005, pp. 12-13.
61. Global Physiological Analysis of Carbon- and Energy- Limited Growing Escherichia coli Confirms a High Degree of Catabolic Flexibility and Preparedness for Mixed Substrate Utilization, J. Ihssen, and T. Egli, Environmental Microbiology, 2005, v.7, pp. 1568-1581.
60. Role of HtrA in Surface Protein Expression and Biofilm Formation by Streptococcus mutants, S. Biswas and I. Biswas, Infection and Immunity, 2005, v.73, pp. 6923-6934.
59. Requirements of the Dephospho-form of Enzyme IIANTR for Derepression of Escherichia coli K-12 ilvBN Expression, C-R. Lee, B-M. Koo, S-H. Cho, Y-J. Kim, M-J. Yoon, A. Peterkofsky, and Y-J. Seok, Molecular Microbiology, 2005, v.58, pp. 334-344.
58. Harnessing Natural Diversity to Probe Metabolic Pathways, O.R. Homann, H. Cai, J.M. Becker, and S.L. Lindquist, PLOS Genetics, 2005, v. 1, pp. 0715-0729.
57. The Phagosomal Transporter A Couples Threonine Acquisition to Differentiation and Replication of Legionella pneumophila in Macrophages, J-D. Sauer, M.A. Bachman, and M.S. Swanson, Proceedings of the National Academy of Sciences, 2005, v.102, pp. 9924-9929.
56. A New Pathway for Pyrimidine Catabolism in E. coli K-12: The b1012-b1006 Operon, K.D. Loh, P. Gyaneshwar, E.M. Papadimitriou, R. Fong, K-S. Kim, Z. Zhou, W. Inwood, and S. Kutsu, Abstract from the International Union of Microbiological Societies, July 2005.
55. Metabolic Analysis of Helicobacter pylori Strain 26695 with Phenotype MicroArray Technology, X-H. Lei, S. Tan, D. E. Berg, and B. R. Bochner, Poster B-760, Session #5-B, presented at International Union of Microbiological Societies, July 2005.
54. Characterization of the Genetic Origins of Phenotypic Diversification of the Pathogenic Salmonellae, B. Bochner, C.A. Morales, S. Porwollik, J.G. Frye, R. Gast, X-H. Lei, M. Ziman, M. McClelland, and J. Guard-Bouldin, Poster B-800, Session # 6-B, presented at International Union of Microbiological Societies, July 2005.
53. Phenotype MicroArray Profiling of Staphylococcus aureus menD and hemB Mutants with Small Colony Variant (SCV) Phenotype, C. von Eiff, P. McNamara, K. Becker, D. Bates, X-H. Lei, M. Ziman, B.R. Bochner, G. Peters, and R.A. Proctor, Poster B-873, Session # 138-B, presented at International Union of Microbiological Societies, July 2005.
52. Use of Phenotype MicroArray Technology to Fingerprint E. coli Loss-of-Function Mutants, M. Ziman, D. Chan, J. Carlson, S. Suharnan, H. Mori, H. Gobara, K. Nakahigashi, T. Baba, S. Tamishima, H. Matsui, and M. Arita, Poster presented at the 7th Annual Frontier of Microbial Research Genome Workshop, March 2005.
51. Proline-Based Modulation of 2,4-Diacetylphloroglucinol and Viable Cell Yields in Cultures of Pseudomonas fluorescens Wild-Type and Over-Producing Strains, P.J. Slininger and M.A. Shea-Andersh, Applied Microbiology and Biotechnology, 2005, v.68, pp. 630-638.
50. Gene Array Analysis of Yersinia enterocolitica FlhD and FlhC: Regulation of Enzymes Affecting Synthesis and Degradation of Carbamoylphosphate, V. Kapatral, J.W. Campbell, S.A. Minnich, N.R. Thomson, P. Matsumura, and B.M. Pruess, Microbiology, 2004, v.150, pp. 2289-2300.
49. Phenotypic MicroArray Provides a Biochemical Profile of Staphylococcus aureus menD and hemB Mutants with Small Colony Variant (SCV) Phenotype, C. von Eiff, P. McNamara, D. Bates, K. Becker, B. Bochner, G. Peters, R.A. Proctor, Poster presented at 11th International Symposium on Staphylococci & Staphylococcal Infections, October 2004.
48. Phenotype MicroArray Analysis of a S. aureus mnhD (NuoN-like) Mutant, a Thrombin-Induced Platelet Microbicidal Protein (tPMP)-Resistant Strain, R.A. Proctor, P.J. McNamara, A. Franco-Buff, B.R. Bochner, J. Schrenzel, M. Yeaman, and A. Bayer, Poster presented at 11th International Symposium on Staphylococci & Staphylococcal Infections, October 2004.
47. Identifying Antimicrobials Using Isobologram Arrays, L.A. Wiater, K. Kim, J.D. Carlson, and P. Gadzinski, Abstract from the 104th General Meeting of the American Society of Microbiology, May 2004.
46. Phenotype MicroArray Analysis of Two-Component Regulatory Genes of Escherichia coli, X-H. Lei, L. Zhou, B.L. Wanner, and B. R. Bochner, Poster K-144, presented at 103rd General Meeting of the American Society of Microbiology, May 2003.
45. Genomic and Physiological Characterization of Desulfovibrio vulgaris Strains Isolated from A Metal Contaminated Lake,S.M. Stolyar, C.B. Walker, H-C. Yen, B. Zuniga, N. Pinel, H. Gough, Z. He, Q. He, J. Zhou, T.C. Hazen, S.E. Borglin, J.D. Wall, and D.A. Stahl, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
44. Phenotype MicroArray Analysis of Desulfovibrio vulgaris, S.E. Borglin, T.C. Hazen, J. Carlson, J. Wall, and D. Joyner, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
43. High Throughput Analysis of Stress Growth Response in Shewanella oneidensis MR1, . Katz, T.C. Hazen, R. Huang, D. Joyner, and S.E. Borglin, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
42. Linkage of 23S and rrlA Single Nucleotide Polymorphisms to Phenotype Heterogeneity of Salmonella Enteritidis, C.A. Morales and J. Guard-Bouldin, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
41. Legionella pneumophilaRequire phtA for Threonine Acquisition, Differentiation and Replication in Murine Bone Marrow Derived Macrophages, J-D. Sauer and M. Swanson, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
40. Phenotype MicroArray analysis of an Escherichia coli O157: H7 luxS Mutant, M. Walters, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
39. Correlation of Phenotype with the Genotype of egg-contaminating Salmonella Enteritidis, C.A. Morales, S. Porwllik, J.G. Frye, H. Kinde, M. McClelland, and J. Guard-Bouldin, Abstract from the ASM 105th Annual Meeting.
38. Only Rarely Does Inorganic Polyphosphate (poly P) Depletion Decrease Motility in Helicobacter pylori, S. Tan, C.D. Fraley, W.L. Beatty, Q. Gong, B.K. Dieckgraefe, A. Kornberg, and D.E. Berg, Abstract from the ASM 105th Annual Meeting, June 5th-June 9th 2005.
37. Correlation of Phenotype with the Genotype of Egg-contaminating Salmonella Enteritidis, C.A. Morales, S. Porwllik, J.G. Frye, H. Kinde, M. McClelland, and J. Guard-Bouldin, Abstract from the ASM Conference on Integrating Metabolism and Genomics, April 30th-May 3rd 2004.
36. Metabolic Differences in Yersinia pestis as a Function of Temperature and Calcium Detected by Phenotype Arrays, A.E. Holtz, I.K. Fodor, J.P. Fitch, and S.L. McCutchen-Maloney, Abstract from the 44th Annual Meeting of the American Society for Cell Biology, December 2004.
35. A Plague on Many Houses, The American Society for Cell Biology Press Book 2004, p 6.
34. LLNLs Ann Holtz-Morris on Studying Y. pestis with Cellular Arrays, Inside BioAssays, Dec. 2004.
33. Probing Microbial Phenotypes in Wake of Genomics Data Flood, J.F. Fox, ASM News, 2000, v.66, pp. 391-392.
32. Characterization of the Escherichia coli AaeAB Efflux Pump: A Metabolic Relief Valve?, T.K. Van Dyk, L.J. Templeton, K.A. Cantera, P.L. Sharpe, and F.S. Sariaslani, Journal of Bacteriology, 2004, v. 186, pp. 7196-7204.
31. Metabolomic Analysis of Escherichia coli K-12 Mutants with Deletions of 1440 Genes, M. Ito, T. Baba, H. Mori, and H. Mori, Abstract from the ASM Integrating Metabolism and Genomics Conference April 30 – May 3, 2004.
30. A Novel Fermentation/Respiration Switch Protein Regulated by Enzyme IIAGlc in Escherichia coli, B.M. Koo, M.J. Yoon, C.R. Lee, T.W. Nam, Y.J. Choe, H. Jaffe, A. Peterkofsky, and Y.J Seok, The Journal of Biological Chemistry, 2004, v.279, pp. 31613-31621.
29. Gene and Bacterial Identification Using High-Throughput Technologies: Genomics, Proteomics, and Phenomics, S.F. Al-Khaldi and M.M. Mossoba, Nutrition, 2004, v.20, pp. 32-38.
28. Integrating High-Throughput and Computational Data Elucidates Bacterial Networks, M.W. Covert, E.M. Knight, J.L. Reed, M.J. Herrgard, and B.O. Palsson, Nature, 2004, v.429, pp. 92-96.
27. Global Analysis of Predicted Proteomes: Functional Adaptation of Physical Properties, C.G. Knight, R. Kassen, H. Hebestreit, and P.B. Rainey, Proceedings of the National Academy of Sciences, 2004, v.101, pp. 8390-8395 .
26. ASAP, a Systematic Annotation Package for Community Analysis of Genomes, J.D. Glasner, P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Brynes, M. Gilson, B. Biehl, F.R. Blattner, and N.T. Perna, Nucleic Acids Research, 2003, v.31, pp. 147-151.
25. Global Nutritional Profiling for Mutant and Chemical Mode-of-Action Analysis in Filamentous Fungi, M.M. Tanzer, H.N. Arst, A.R. Skalchunes, M. Coffin, B.A. Darveaux, R.W. Heiniger, and J.R. Shuster, Functional Integrated Genomics, 2003, v.3, pp. 160-170.
24. Fast Tracking Bioprocessing Development and Optimisation, C. Gorman, Drug Plus International, 2003, v.2 pp. 6-9.
23. Phenotype MicroArray Analysis of Escherichia coli K-12 Mutants with Deletions of All Two-Component Systems, L.Zhou, X.-H. Lei, B.R. Bochner, and B.L. Wanner, Journal of Bacteriology, 2003, v.185, pp. 4956-4972.
22. Experimental Adaptive Radiation in Pseudomonas, R.C. MacLean and G. Bell, The American Naturalist, 2002, v.160 pp. 569-581.
21. Divergent Evolution During an Experimental Adaptive Radiation, R.C. MacLean and G. Bell, Proc. Biol. Sci., 2003, v.270 pp. 1645-1650.
20. New Technologies to Assess Genotype-Phenotype Relationships, B.R. Bochner, Nature Reviews Genetics, 2003, v.4 pp. 309-314.
19. Phenotype MicroArrays: Their Use in Antibiotic Discovery, B.R. Bochner, Chapter 9 in Microbial Genomics and Drug Discovery, edited by T.J. Dougherty and S.J. Projan, Marcel Dekker, Inc., 2003.
18. Phenotype MicroArrays for High-Throughput Phenotypic Testing and Assay of Gene Function, B.R. Bochner, P. Gadzinski, and E. Panomitros, Genome Research, 2001, v.11, pp. 1246-1255.
17. Identification of Bacteria and Bacterial Genes: Infrared Spectroscopy, Genomics, Proteomics and Phenomics, S. Al-Khaldi and M. Mossoba, Microbiological Method Forum News, 2003, v.23, pp.1-6.
16. FlhD/FlhC is a Regulator of Anaerobic Respiration and the Entner-Doudoroff Pathway through Induction of the Methyl-Accepting Chemotaxis Protein Aer, B.M. Prub, J.W. Campbell, T.K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura, Journal of Bacteriology, 2003, v.185, pp. 534-543.
15. Next-Generation Microarray Technologies, B.J. Sedlack, Genetic Engineering News, 2003, v.23, p.20.
14. Engineering a Reduced Escherichia coli Genome, V. Kolisnychenko, G. Plunkett, III, C.D. Herring, T. Feher, J. Posfai, F.R. Blattner, and G. Posfai, Genome Research, 2002, v.12, pp. 640-647.
13. Carbon and Nitrogen Substrate Utilization by Archival Salmonella typhimurium LT2 Cells, B.S. Tracy, K.K. Edwards, and A. Eisenstark, BMC Evolutionary Biology, 2002, v.2, pp. 1-6.
12. Regulation of Carbon Utilization by Sulfur Availability in Escherichia coli and Salmonella typhimurium, J.A. Quan, B.L. Schneider, I.T. Paulsen, M. Yamada, N.M. Kredich, and M.H. Saier, Jr., Microbiology, 2002, v.148, pp. 123-131.
11. Gene Discovery and Gene Function Assignment in Filamentous Fungi, L. Hamer, K. Adachi, M.V. Montenegro-Chamorro, M.M. Tanzer, S.K. Mahanty, C. Lo, R.W. Tarpey, A.R. Skalchunes, R.W. Heiniger, S.A. Frank, B.A. Darveaux, D.J. Lampe, T.M. Slater, L. Ramamurthy, T.M. DeZwaan, G.H. Nelson, J.R. Shuster, J. Woessner, and J.E. Hamer, Proc. Natl. Acad. Sci. USA, 2001, v.9, pp. 5110-5115.
10. Discovery Technologies, Intermediate Steps Follow Up On Genome Project, N. Flanagan, Genetic Engineering News, 2001, v.21, p.1.
9. Proteome Chips Promise to Speed Decoding of the Human Genome, M. Downs, Small Times, 2001.
8. Phenotyping Goes High Throughput with Biolog Phenotype MicroArrays, J. Lane, The Scientist, 2001, v.15.
7. High-Throughput Cell Analytic Tool, B.R. Bochner, Drug Discovery/Technology News, 2001, v.4.
6. Phenotype Arrays, A. Marshall, Nature Biotechnology, 2001, v.19, pp. 629.
5. Cellular Assays to Drug Discovery, T. Mullane, Pharmaceutical Laboratory, June 2000.
4. The Population Genetics of Ecological Specialization in Evolving Escherichia coli Populations, V. S. Cooper and R. E. Lenski, Nature, 2000, v.407, pp. 736-739.
3. Megaplasmid pRme2011a of Sinorhizobium meliloti is Not Required for Viability, I. J. Oresnik, S. Liu, C. K. Yost, and M. F. Hynes, Journal of Bacteriology, 2000, v.182, pp. 3582-3586.
2. The Consequences of Growth of a Mutator Strain of Escherichia coli as Measured by Loss of Function Among Multiple Gene Targets and Loss of Fitness, P. Funchain, A. Yeung, J. L. Stewart, R. Lin, M. M. Slupska, and J. H. Miller, Genetics Society of America, 2000, v.154, pp. 959-970.
1. Diagnostic Advances: Genotypic, Phenotypic, Fast, J.F. Fox, ASM News, 2000, v.66, pp. 391-392.
- :Omnilog PM
- :美国Biolog
- :Omnilog PM可用来测定微生物细胞的表型,是与DNA芯片和蛋白质组技术并列的、应用在 基因组研究和药物开发的三大技术之一。配套25块PM板,其中碳源板2块、磷酸源板1块,氨基酸和短肽板4块、生物合成路径板1块、离子强度板1块、pH板1块、抗生素板15块(其中细菌10块,真菌5块)。用于微生物代谢研究、基因功能研究、发酵条件优化和产生形成条件优化、耐药研究等。


